« Prev Next »
Every living organism must be able to sense environmental stimuli and convert these input signals into appropriate cellular responses. Most of these responses are mediated by transcription factors that bind DNA and coordinate the activity of RNA polymerase or of proteins that elicit allosteric effects on their regulatory targets. In the early 1970s, researchers began to recognize that regions of mRNA transcripts have a regulatory role in the expression of downstream gene products (Jackson & Yanofsky 1973). By the early 1990s, several new regulatory mechanisms had been discovered that center on the action of RNA. (Arnaud et al. 1996; Aymerich & Steinmetz 1992; Houman et al. 1990; Lu et al. 1996; Oda et al. 2000; Wray & Fisher 1994). One classic example of regulation by RNA (often referred to as riboregulation) was discovered by Charles Yanofsky, who described the regulation of tryptophan biosynthesis at the mRNA level through the coupling of translation and transcription (Babtizke & Yanofsky 1993; Otridge & Gollnick 1993; Shimotsu et al. 1986; Yanofsky et al. 1996). Since then, many diverse RNA-based regulatory mechanisms have been discovered, including one that regulates interference and epigenetic regulation by long, noncoding RNA in eukaryotes (Costa 2007; Mattick 2001).
Riboregulation in Bacteria
| Table 1: Representative riboswitch classes A broad overview of known riboswitches, including the classes that are the best understood to date. | ||||
| Ligand type | Ligand name | Riboswitch family/class | Discovery (year) | Structure (year) |
| Enzyme cofactor | S-adenosylmethionine | SAM/SAM-I | 2003 (Winkler et al. 2003) | 2006 (Montange & Batey 2006) |
| S-adenosylmethionine | SAM/SAM-II | 2005 (Corbino et al. 2005) | 2008 (Gilbert et al. 2008) | |
| S-adenosylmethionine | SAM/SAM-III | 2006 (Fuchs et al. 2006) | 2008 (Lu et al. 2008) | |
| Thiamine pyrophosphate | TPP | 2001 (Miranda-Rios et al. 2001) | 2006 (Serganov et al. 2006; Thore et al. 2006) | |
| Nucleotide precursor | Guanine | Purine/G | 2003 (Mandal et al. 2003) | 2004 (Batey et al. 2004; Serganov et al. 2004) |
| Adenine | Purine/A | 2003 (Mandal & Breaker 2004) | 2004 (Serganov et al. 2004) | |
| 2'-Deoxyguanosine | Purine/dG | 2007 (Kim et al. 2007) | 2009 (Edwards & Batey 2009) | |
| Amino acid | Lysine | Lysine | 2003 (Rodionov et al. 2003; Sudarsan et al. 2003) | 2008 (Garst et al. 2008; Serganov et al. 2008) |
| Metal ion | Magnesium | Mg2+/ykoK | Identified 2004 (Barrick et al. 2004); validated 2007 (Dann et al. 2007) | 2007 (Dann et al. 2007) |
What Is the Structure of a Riboswitch?
Riboswitches are composed of two domains: the aptamer domain and the expression platform (Figure 1; Tucker & Breaker 2005). The aptamer domain acts as a receptor that specifically binds a ligand (Figure 1, red). The expression platform acts directly on gene expression through its ability to toggle between two different secondary structures in response to ligand binding (Figure 1, blue). Common to both domains is something called the switching sequence, and its placement in the aptamer domain or the expression platform ultimately dictates the expression outcome of the mRNA (Figure 1, purple). Specifically, if metabolite binding to the riboswitch stabilizes incorporation of the switching sequence into the aptamer domain, the expression platform must fold into a specific structure. Riboswitches that control transcriptional repression have a switching sequence that directs formation of a Rho-independent transcriptional terminator, a short stem-loop structure (followed by six or more uridine residues) that signals RNA polymerase to abort transcription (Nudler & Gottesman 2002; Hammann & Westhof 2007). Other riboswitches that regulate translational initiation utilize a switching sequence that can expose or occlude a ribosome-binding site (called the Shine-Dalgarno sequence; Hammann & Westhof 2007).
How Are Riboswitches Categorized?
Riboswitches are organized into families and classes according to two features: the type of ligand they bind, and their secondary structure (the arrangement of Watson-Crick paired helices; Hammann & Westhof 2007; Montange & Batey 2008). A family of riboswitches is typically a group of RNAs related by the ligands they recognize. For example, the SAM riboswitch family recognizes the compound S-adenosylmethionine (SAM). Within a family, there may be distinct classes of riboswitches, each class distinguished by a common sequence pattern that usually defines the ligand-binding pocket, as well as features required for folding the RNA into a three-dimensional shape. The SAM riboswitch family contains at least five known classes (Wang & Breaker 2008). These classes are distinguished from one another by their architectural features. For example, the SAM-I class forms a four-way helical junction, SAM-II forms a classic (H-type) pseudoknot, and SAM-III is defined by a three-way junction (Figure 3; Montange & Batey 2006; Gilbert et al. 2008; Lu et al. 2008).

Purine Riboswitch Family: Global Structure
An exception to the classification scheme described above is the purine riboswitch family, a group of RNAs that actually share a common secondary structure but can recognize multiple distinct ligands (Kim & Breaker 2008).
The purine riboswitch family includes the adenine, guanine, and 2'-deoxyguanosine classes. Because its members recognize multiple ligands, this riboswitch family serves as a model for understanding the mechanisms of ligand recognition (Kim & Breaker 2008). The global architecture of the RNA in a purine riboswitch is defined by the organization of the three conserved helices that make up the secondary structure (Figure 4A). Two of the RNA helices form a coaxial stack, meaning that one helix sits on top of the other, and they are collinear. This pairing is the basis for their names, P1 and P3; P is an abbreviation for "paired" (Figure 4B). The third helix (P2) is adjacent to P3, and the terminal loops of P2 and P3 together form a tertiary structure called a loop-loop. (Because they form loops, P2 is also called L2, and P3 is also called L3 — since L is an abbreviation for "loop"). A complex set of interacting helices and loop formations defines the overall three-dimensional fold of the purine riboswitch aptamer domain where ligands bind (Batey et al. 2004; Serganov et al. 2004).
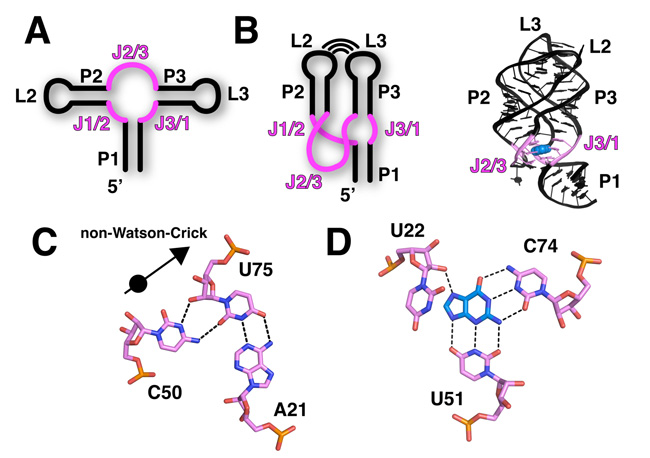
Purine Riboswitch Family: Ligand Binding in the Aptamer Domain
The three-way helical junction where P1, P2, and P3 meet is the ligand-binding pocket of a purine riboswitch. This region of the RNA is defined by a series of noncanonical base interactions (i.e., interactions that are not exclusively involved in Watson-Crick pairing). For example, in the P1 helix proximal to the ligand-binding site, a base triple interaction is observed in most purine riboswitches (Figure 4C). This triple, as well as most other unusual base triples, is typically composed of a Watson-Crick pair (A21-U75) interacting with a third base (C50). At the center of the junction, a pyrimidine (Y) at position 74 forms a Watson-Crick pairing interaction with the ligand, which is further surrounded by other conserved residues (Figure 4D). The identity of this pyrimidine residue (cytosine or uridine) is the basis for specificity between the guanine and adenine classes (Noeske et al. 2005; Gilbert et al. 2006).
One of the most notable features of the ligand-bound aptamer domain is the way it nearly completely encapsulates the ligand, with the surrounding aqueous environment making the ligand almost entirely inaccessible (Batey et al. 2004; Serganov et al. 2004). Scientists have observed this encapsulation in other riboswitches, suggesting that this sequestering of the ligand is an important feature for the riboswitch mechanism of gene regulation (Edwards et al. 2007). The observation of this commonality also implies that ligand binding is often accompanied by a local conformational change in the aptamer domain, such that one element closes down on the binding pocket like a lid. What mechanism controls this lid closing? Scientists have used a combination of chemical probing experiments, fluorescence-based measurements, and kinetic analyses to look for an answer. Here is what some have proposed: one strand of the three-way junction, J3/1 (J for "joining") acts as a preformed docking station for the incoming purine ligand by making Y74 available for Watson-Crick pairing. Once the ligand is paired with Y74, nucleotides comprising J2/3 then fold over the bound ligand and encapsulate it (Gilbert et al. 2006; Noeske et al. 2007; Stoddard et al. 2008).
Purine Riboswitch Family: Ligand Binding and Regulatory Mechanism
The coupling of ligand binding to a conformational change is central to the regulatory mechanism of the riboswitch. As J2/3 encapsulates the ligand, it also forms additional tertiary interactions with the 3' strand of the P1 helix (including the base triple shown in Figure 4D). This region of the P1 helix is the switching sequence (Figure 1, purple). These additional ligand-induced interactions with the P1 helix stabilize its incorporation into the aptamer domain. Why is this important? It prevents the helix from being utilized to form alternative structures in the expression platform. In this way, the expression platform is fated to form one of two structures that interface with the expression machinery: either RNA polymerase or the ribosome. Thus, the ligand-binding event and the downstream regulatory switch are coupled through a limited ligand-induced conformational change that involves the switching sequence. This concept, originally proposed after scientists figured out the structure of the first riboswitch, has proven to be a nearly universal feature of riboswitches (Batey et al. 2004).
Kinetic Control of Riboswitches: A Race with Transcription
Given these observations about ligand concentration, ligand-riboswitch binding, and riboswitch activation, scientists have proposed a more plausible model, stating that riboswitches are under kinetic control (Wickiser, Cheah, et al. 2005; Wickiser, Winkler, et al. 2005; Gilbert et al. 2006). Recall that a kinetically controlled event is one that depends largely or entirely on the rate-limiting step of a chemical process. For riboswitches, we must understand the relationship between the rates of transcription, RNA folding, and ligand binding. During transcription, the aptamer domain is always the first part of the RNA to be synthesized and fold into a shape capable of binding ligand. After completion of the aptamer domain, often a programmed pause site in the mRNA causes the polymerase to temporarily stall (Figure 5, star). This short pause (actually measured to be 2–10 seconds) gives the aptamer domain time to "interrogate" the cellular environment for the presence of ligand (Wickiser, Winkler, et al. 2005). If a sufficient concentration of ligand is present, then the ligand will occupy the binding pocket prior to resumption of transcription, and cause the mRNA to form a Rho-independent transcriptional terminator. Often a second pause site midway through the expression platform gives the RNA time, if ligand has not bound, to reconfigure the RNA secondary structure. In this case, the switching sequence will be used to form the antiterminator (Figure 5, AT), a stem-loop structure that disallows formation of the terminator, resulting in full transcription of the mRNA and allowing for its expression into protein.
The kinetic control component of this process is the balance of rates of several processes: (1) speed of riboswitch transcription, including time spent residing at any potential pause sites; (2) ligand binding; and (3) a possible secondary structural rearrangement. Because several studies have shown that the rate of ligand binding to the aptamer domain is slow, the cellular concentration of ligand must be far greater than the dissociation constant (KD) in order for binding to be sufficiently rapid to beat the kinetics of transcription (Wickiser, Cheah, et al. 2005; Wickiser, Winkler, et al. 2005; Gilbert et al. 2006).
How Riboswitches Can Affect Humans
Since these fascinating riboswitches are mechanisms specific to bacteria, it may be difficult determine how relevant they are to humans and human health. However, their role in regulating transcription in bacteria makes them enticing targets for the development of novel antibiotics aimed at stopping bacterial pathogens from flourishing inside the people they infect. Because riboswitches control genes essential for bacterial survival, or genes that control the ability of bacteria to succeed at infection, a drug designed to affect a riboswitch could be a powerful tool for shutting down pathogenic bacteria (Blount & Breaker 2006). In fact, many antimicrobial compounds affect RNA directly, and many commonly used antibiotics inhibit translation by targeting bacterial ribosomes through binding interactions with ribosomal RNA (Vicens & Westhof 2003; Sutcliffe 2005). In addition, some compounds bind to the lysine, TPP, and FMN riboswitch classes and slow bacterial cell growth (Sudarsan et al. 2005; Blount et al. 2007; Lee et al. 2009).
Optimism for Novel Riboswitch-Based Therapeutics
There are two main reasons to expect that drugs designed to inhibit riboswitches would have minimal side effects in humans. One is that riboswitches have not been identified in mammals, so they are not likely to act on mammalian mRNA. Another is that some riboswitches are known to bind their cognate ligand in fundamentally different ways than do mammalian proteins that recognize the same ligand (Montange & Batey 2006). This means there is reason to suspect they won't interfere with ligand binding in native mammalian systems. Scientists have made some advances toward this kind of riboswitch-based drug design by figuring out the details of riboswitch-ligand complex structure. These structural data provide the basis for designing drugs that interfere with the riboswitch complexes and affect their shape, the fundamental basis for riboswitch control of RNA transcription.
References and Recommended Reading
Arnaud, M. et al. In vitro reconstitution of transcriptional antitermination by the SacT and SacY proteins of Bacillus subtilis. Journal of Biological Chemistry 271, 18966–18972 (1996)
Aymerich, S. & Steinmetz, M. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. PNAS 89, 10410–10414 (1992)
Babitzke, P. & Yanofsky, C. PNAS 90, 133–137 (1993)
Batey, R. T., Gilbert, S. D. & Montange, R. K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432, 411–415 (2004)
Blount, K. F. & Breaker, R. R. Riboswitches as antibacterial drug targets. Nature Biotechnology 24, 1558–1564 (2006)
Blount, K. F. et al. Antibacterial lysine analogs that target lysine riboswitches. Nature Chemical Biology 3, 44–49 (2007)
Costa, F. F. Non-coding RNAs: Lost in translation? Gene 386, 1–10 (2007)
Dann, C. E. et al. Structure and mechanism of a metal-sensing regulatory RNA. Cell 130, 878–892 (2007)
Edwards, T. E., Klein, D. J. & Ferre-D'Amare, A. R. Riboswitches: Small-molecule recognition by gene regulatory RNAs. Current Opinion in Structural Biology 17, 273–279 (2007)
Gilbert, S. D. et al. Structure of the SAM-II riboswitch bound to S -adenosylmethionine. Nature Structural and Molecular Biology 15, 177–182 (2008)
Gilbert, S. D. et al. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. Journal of Molecular Biology 359, 754–768 (2006)
Hammann, C. & Westhof, E. Searching genomes for ribozymes and riboswitches. Genome Biology 8, 210 (2007)
Houman, F., Diaz-Torres, M. R. & Wright, A. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell 62, 1153–1163 (1990)
Jackson, E. N. & Yanofsky, C. Thr region between the operator and first structural gene of the tryptophan operon of may have a regulatory function. Journal of Molecular Biology 76, 89–101 (1973)
Kim, J. N. & Breaker, R. R. Purine sensing by riboswitches. Biology of the Cell 100, 1–11 (2008)
Lee, E. R., Blount, K. F. & Breaker, R. R. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biology 6, 187–194 (2009)
Lu, C. et al. Crystal structures of the SAM-III/S(MK) riboswitch reveal the SAM-dependent translation inhibition mechanism. Nature Structural and Molecular Biology 15, 1076–1083 (2008)
Lu, Y., Turner, R. J. & Switzer, R. L. Function of RNA secondary structures in transcriptional attenuation of the operon. PNAS 93, 14462–14467 (1996)
Mattick, J. S. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Reports 2, 986–991 (2001)
Montange, R. K. & Batey, R. T. Riboswitches: Emerging themes in RNA structure and function. Annual Reviews of Biophysics 37, 117–133 (2008)
Montange, R. K. & Batey, R. T. Structure of the S -adenosylmethionine riboswitch regulatory mRNA element. Nature 441, 1172–1175 (2006)
Noeske, J. et al. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. PNAS 102, 1372–1377 (2005)
Noeske, J. et al. Interplay of "induced fit" and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Research 35, 572–583 (2007)
Nudler, E. & Gottesman, M. E. Transcription termination and anti-termination in E. coli. Genes to Cells 7, 755–768 (2002)
Nudler, E. & Mironov, A. S. The riboswitch control of bacterial metabolism. Trends in Biochemical Sciences 29, 11–17 (2004)
Oda, M. et al. operon and histidine-dependent binding of HutP to the transcript containing the regulatory sequences. Molecular Microbiology 35, 1244–1254 (2000)
Otridge, J. & Gollnick, P. MtrB from RNA in a tryptophan-dependent manner. PNAS 90, 128–132 (1993)
Serganov, A. et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chemistry and Biology 11, 1729–1741 (2004)
Shimotsu, H. et al. Novel form of transcription attenuation regulates expression the tryptophan operon. Journal of Bacteriology 166, 461–471 (1986)
Stoddard, C. D., Gilbert, S. D. & Batey, R. T. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA 14, 675–684 (2008)
Sudarsan, N. et al. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chemistry and Biology 12, 1325–1335 (2005)
Sutcliffe, J. A. Improving on nature: Antibiotics that target the ribosome. Current Opinion in Microbiology 8, 534–542 (2005)
Tucker, B. J. & Breaker, R. R. Riboswitches as versatile gene control elements. Current Opinion in Structural Biology 15, 342–348 (2005)
Vicens, Q. & Westhof, E. RNA as a drug target: The case of aminoglycosides. Chembiochem 4, 1018–1023 (2003)
Wachter, A. et al. Riboswitch control of gene expression in plants by splicing and alternative 3' end processing of mRNAs. Plant Cell 19, 3437–3450 (2007)
Wang, J. X. & Breaker, R. R. Riboswitches that sense . Biochemistry and Cell Biology 86, 157–168 (2008)
et al. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry 44, 13404–13414 (2005)
Wickiser, J. K., Winkler, W. C. et al. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Molecular Cell 18, 49–60 (2005)
Winkler, W. C. Riboswitches and the role of noncoding RNAs in bacterial metabolic control. Current Opinion in Chemical Biology 9, 594–602 (2005)
Wray, L. V., Jr. & Fisher, S. H. Analysis of Bacillus subtilis hut operon expression indicates that histidine-dependent induction is mediated primarily by transcriptional antitermination and that amino acid repression is mediated by two mechanisms: Regulation of transcription initiation and inhibition of histidine transport. Journal of Bacteriology 176, 5466–5473 (1994)
Yanofsky, C., Konan, K. V. & Sarsero, J. P. Some novel transcription attenuation mechanisms used by bacteria. Biochimie 78, 1017–1024 (1996)



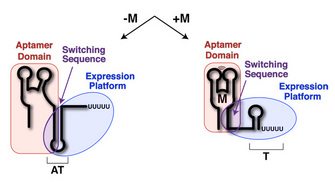
 Figure 1: Riboswitch domains
Figure 1: Riboswitch domains




























