« Prev Next »
DNA replication occurs during the synthesis (S) phase of the cell cycle and starts at predefined DNA sequences known as replication origins; this start is also known as origin firing (Errico & Costanzo 2010; Koren et al. 2010). Once the origins of replication have fired, the DNA replication proteins organize into a structure called the replication fork (RF), where a group of proteins coordinate DNA replication (Langston et al. 2009). It is called a fork because the simplified structure resembles a two-tined fork, but its function is hardly simple, since it is the location of dynamic activity among a large group of proteins (Figure 1).
What Happens at the Replication Fork?
Two main activities happen at the fork: DNA unwinding and DNA synthesis. The RF unwinds the unreplicated DNA ahead of it through a helicase enzyme complex (Gambus et al. 2009; Lou et al. 2008). As their name suggests, helicases modify the structure of the DNA helix and promote unwinding and separation of the two DNA strands. The second activity of DNA synthesis at the RF is undertaken by DNA polymerase. This enzyme links together, or polymerizes, DNA bases in the correct sequence using the template DNA strand, and it generates two copies of the genome that are later divided into daughter cells in metaphase, or M phase (Langston et al. 2009).
How Does a Replication Fork Stall?
A variety of impediments can stall the RF (Labib & Hodgson 2007; Rothstein et al. 2000; Torres-Rosell et al. 2007). Functionally this means that the RF complex is paused, sometimes indefinitely, before restarting. The cell's response to RF stalling depends on the number of stalled forks and the length of the arrest (Labib & Hodgson 2007; Lucca et al. 2004; Meister et al. 2005; Tourriere & Pasero 2007). By studying how RFs arrest and restart in the laboratory, we can learn how defects in RF function can damage cells. Some chemicals can cause RF-stalling DNA damage that leads to diseases such as cancer (Bartkova et al. 2005; Calzada et al.,2005; Wray et al. 2008). Therefore scientists study these drug effects on RF arrest in model organisms to understand the effect on DNA mutagenesis and cell viability.
Scientists study stalled RFs in the laboratory using the drug hydroxyurea (HU). HU causes the depletion of deoxynucleotide triphosphates (dNTPs) in cells (Bianchi et al. 1986; Koc et al. 2004; Matsumoto et al. 1990). DNA polymerases use dNTPs as building material for DNA synthesis and elongation. Without dNTPs, the polymerase component of RFs cannot continue and slows down and arrests (Feng et al. 2006; Mirkin & Mirkin 2007).
Because polymerase stalling causes RF complex arrest, these RF activities are linked (Figure 2). In particular, the helicase-initiated unwinding activity that precedes the RF is functionally linked to the polymerization activity (Calzada et al. 2005; Labib 2008; Lou et al. 2008; Tanaka, Katou et al. 2009). To overcome RF stalling, the cell may adapt to or overcome HU inhibition, or it may be released from stalling when HU is removed from the environment and the dNTP supply is replenished in the cell (Kurose et al. 2006; Lopes et al. 2001; Mulder et al. 2005). Irrespective of how long the RF is arrested to make necessary repairs, RFs must be capable of restarting so that the cell cycle can finish. To further complicate matters, cells with stalled RFs are at an increased risk for DNA damage that can lead to mutation or cell death (Bernstein et al. 2009; Bryant et al. 2009; Froget et al. 2008; Kai et al. 2005; Mao et al. 2009; Noguchi et al. 2003; Petermann et al. 2010). Thus, cells have distinct challenges in the response to and recovery from RF stalling and can be studied using HU in laboratory experiments. How are stalled RFs stabilized so that replication can resume?
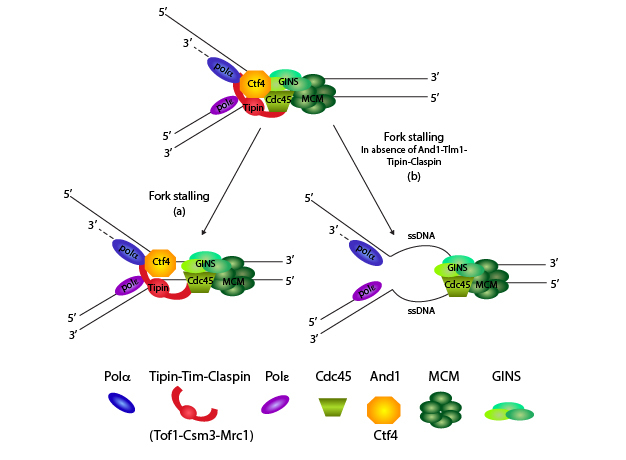
Setting the Stage: Considering Replication Fork Activity
When RFs stall upon dNTP depletion, how does the cell detect this problem and respond? The RF helicase is a subcomplex of the fork complex itself, and is made up of the hexameric minichromosome maintenance (MCM) helicase and several additional factors (see Figure 1; Gambus et al. 2009; Nedelcheva et al. 2005; Tanaka, Katou et al. 2009). The MCM helicase travels with the RF, unwinding DNA ahead of the RF to make single-stranded DNA (ssDNA) available (Calzada et al. 2005; Langston et al. 2009; Nedelcheva et al. 2005). ssDNA is the substrate for both leading- (Pole) and lagging-strand (Pold and Pola-primase) polymerases. The ssDNA becomes coated with replication protein A (RPA), the ssDNA-binding protein that ensures that the two separated DNA strands remain separated.
The MCM unwinding activity must remain linked to the polymerization activity to prevent excessive unwinding, which could also cause DNA damage (Gambus et al. 2009; Nedelcheva et al. 2005; Razidlo & Lahue 2008). If polymerization stops, the helicase has to stop as well. A candidate complex to maintain this linkage is the fork protection complex (FPC), which contains four conserved proteins: Timeless, Tipin, Claspin and And1 (Figure 2; Kemp et al. 2010; Lou et al. 2008; Noguchi & Noguchi 2007; Noguchi et al. 2004; Unsal-Kacmaz et al. 2007; Yoshizawa-Sugata & Masai 2007). These fork stability components affect not only proper RF function and the ability to stall, but also the ability of chromosomes to segregate properly in metaphase
Timeless and Tipin Contribute to RF Replication and Stability
Timeless and Tipin (derived from the TIM and TIPIN genes) interact with other components of the RF and form the core of the FPC (Table 1 and Figure 2). These proteins have evolutionarily conserved roles in DNA replication and RF stability (Errico et al. 2007; Gotter et al. 2007; Katou et al. 2003; McFarlane et al. 2010; Noguchi et al. 2003, 2004). In both yeast and humans, Timeless and Tipin complex (also Tim/Tipin) promotes RF activity and is particularly important in DNA with repetitive sequences that are error prone (Dalgaard & Klar 2000; Krings & Bastia 2004; Noguchi & Noguchi 2007; Razidlo & Lahue 2008). These naturally occurring DNA sequences that are at risk for pausing the RF even in the absence of external factors (such as HU) emphasize the importance of RF stalling. Although not essential for DNA replication, Tim/Tipin promote promotes RF stability (Calzada et al. 2005; Errico et al. 2009; Gambus et al. 2006; Katou et al. 2003; Nedelcheva et al. 2005). Thus, these FPC proteins contribute to cell viability and mutational avoidance in undisturbed and deliberately stalled replication.
| Table 1: Fork protection complex components and homologs from yeast to metazoa. The proteins of the FPC are conserved from yeast to higher eukaryotes. Although the names are different between model organisms, the function of each component within the FPC is similar. In addition, removal of a given component often elicits the same phenotype; for example, loss of And1 (in frog, Xenopus laevis) or Ctf4 (in yeast) affects chromosome cohesion. | |||
| Replication fork component (human) | Protein homologs In other species | Role in FPC | References |
| TIM1 |
Tim1 (Mouse, Xenopus laevis) Tof1 (S. cerevisiae) Swi1 (S. pombe) |
Fork stability; affects sister chromatid cohesion; maintains genome stability | Gotter et al., 2007; Leman et al., 2010; McFarlane et al., 2010; Urtishak et al., 2009 |
| TIPIN |
Tipin (Mouse, Xenopus laevis) Csm3 (S. cerevisiae) Swi3 (S. pombe) |
Fork stability; has little effect on DNA replication if removed; affects sister chromatid cohesion | Errico et al., 2007; Gotter et al., 2007; Kemp et al., 2010; Leman et al., 2010; Smith et al., 2009; Yoshizawa-Sugata & Masai, 2007 |
| CLASPIN |
Claspin (Mouse, Xenopus laevis) Mrc1 (S. cerevisiae, S. pombe) |
Connects helicase and polymerase epsilon; plays role in RF stability; involved in checkpoint signaling; interacts directly with CHK1/RAD53/CDS1 effector kinases, as well as MUS81 nuclease | Alcasabas et al., 2001; Calzada et al., 2005; Komata et al., 2009; Koren et al., 2010; Lou et al., 2008; Naylor et al., 2009; Nedelcheva et al., 2005; Osborn & Elledge, 2003; Szyjka et al., 2005; Tourriere et al., 2005; Zhao et al., 2003 |
| AND1 |
AND1 (Mouse, Xenopus laevis) CTF4 (S. cerevisiae) MCL1 (S. pombe) |
Connects helicase and polymerase alpha; has some effect on DNA replication, magnified with TIPIN depletion; loss affects chromatid cohesion and segregation; plays role in genome stability | Bando et al., 2009; Errico et al., 2009; Gambus et al., 2009; Hanna et al., 2001; Im et al., 2009; Mamnun et al., 2006; Tanaka et al., 2009; Williams & McIntosh, 2002; Zhu et al., 2007 |
The FPC also assists in transmitting signals of RF stalling that help coordinate other steps in the cell cycle (Figure 3; McFarlane et al. 2010; Unsal-Kacmaz et al. 2007; Yoshizawa-Sugata & Masai 2007). For example, Tim/Tipin interacts with cohesin, the protein that holds sister chromatids together. Removal or disruption of Tim/Tipin disturbs sister chromatid cohesion, resulting in more space between chromatids (Errico et al. 2009; Leman et al. 2010; Tanaka, Kubota et al. 2009). In these ways, Tim/Tipin influences DNA replication, fork stalling and the response to stalling, and proper chromosome segregation in metaphase.
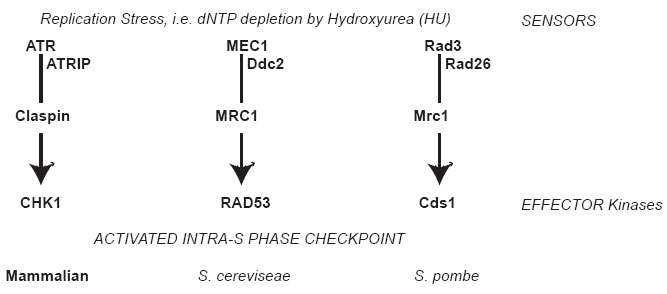
Claspin Maintains Replication Fork Speed and Efficiency
Claspin is another component of the FPC that is involved in multiple stages of DNA replication, particularly uninterrupted replication. We know a great deal about its function from studying the yeast strains that lack the Claspin homolog Mrc1 (∆mrc1 mutants; Osborn & Elledge 2003). Interestingly, ∆mrc1 cells exhibit increased dormant origin firing (Koren et al. 2010), demonstrating the role of Mrc1 in regulating the start of replication. In addition, ∆mrc1 cells replicate DNA more slowly than wild type cells in unstressed conditions (Szyjka et al. 2005), suggesting that Mrc1 function is important for normal replication speed and efficiency. Mrc1 associates with Tof1/Csm3 (Tim/Tipin homolog) linking the leading-strand polymerase, Pole, to the helicase (Figure 2; Bando et al. 2009; Katou et al. 2003).
Mrc1 also conveys the ssDNA-RPA signal created by stalled RFs to downstream signaling molecules and initiates the intra-S phase checkpoint (Tanaka & Russell 2001, 2004; Zhao et al. 2003). For example, Xu and colleagues found that fission yeast Mrc1 is phosphorylated to recruit the effector kinase (Cds1) to the stalled RF, allowing Cds1 to become active and stabilizing the stalled RF (Figure 3; Xu et al. 2006). They demonstrated that Cds1 recruitment through Mrc1 (first step) is essential for Cds1 activation and signal transmission (second step). This two-step model relies on both the presence of Mrc1 at the stalled RF and Mrc1 modification (phosphorylation) by the upstream kinase Rad3, in response to RF stalling (Xu et al. 2006). This signal transduction via Mrc1, and the importance of Mrc1 at the RF, is similar in Saccharomyces cerevisiae (Alcasabas et al. 2001; Branzei & Foiani 2006, 2007; Naylor et al. 2009; Osborn et al. 2002; Schleker et al. 2009; Tourriere & Pasero 2007). Therefore Mrc1 is a replication-specific mediator protein that brings together signaling components to transmit the signal of RF stalling (Alcasabas et al. 2001; Osborn et al. 2002).
However, Mrc1 modification/phosphorylation is also important for its interactions with other RF components. Lou et al. (2008) discovered that Mrc1 phosphorylation changes its interaction with Polε at the RF. Hence, the Mrc1 and polymerase interaction is flexible (Lou et al. 2008). During RF stalling caused by HU, does Mrc1 phosphorylation alter the Mrc1-RF interaction and allow remodeling of other components at the RF?
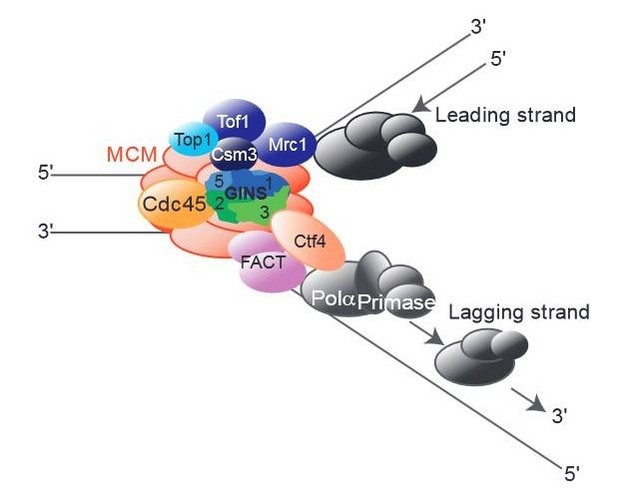
The Role of And1
The last of the FPC proteins is And1, which links the lagging-strand primase (Pola) with helicase function (Figures 4 and 5; Gambus et al. 2009; Im et al. 2009; Miles & Formosa 1992; Williams & McIntosh 2002). The yeast homologs are Ctf4 and Mcl1 (Table 1; Miles & Formosa 1992; Williams & McIntosh 2002). And1 stabilizes Polα association with the RF, and it is important for normal DNA replication (Figure 4; Errico et al. 2009; Tanaka, Katou et al. 2009; Tanaka, Kubota et al. 2009; Zhu et al. 2007). In yeast, the homolog Ctf4 links the helicase to Polα (Figure 5; Gambus et al. 2009).
This And1/Ctf4/Mcl1 trio also influences chromosome segregation by promoting proper chromatid cohesion and centromere assembly prior to metaphase (Hanna et al. 2001; Mamnun et al. 2006; Tanaka, Kubota et al. 2009; Williams & McIntosh 2002). And1 function in RF activity and stability, coupled with its requirement to ensure proper chromosome segregation, highlights the importance of the FPC to later cell cycle events.

Summary
HU-stalled RFs are stabilized by the presence of the FPC, which includes Tim/Tipin, Clapsin, and And1 (Calzada et al. 2005; Noguchi et al. 2003; Tourriere & Pasero 2007). These proteins link helicase and polymerase activities, ensuring proper DNA replication and chromosome segregation (Gambus et al. 2006; Leman et al. 2010; Lopes et al. 2001; Szyjka et al. 2005; Tanaka, Kubota et al. 2009; Urtishak et al. 2009; Yoshizawa-Sugata & Masai 2009). The FPC provides a platform for damage response signaling and mediates interactions with kinases that trigger an intra-S phase arrest (Boddy et al. 1998; Branzei & Foiani 2007; Brondello et al. 1999; Kumagai et al. 2004; Lee et al. 2005; Unsal-Kacmaz et al. 2007; Yoshizawa-Sugata & Masai 2007). Research into the functions of these proteins enables us to understand how replication proceeds normally, and what mechanisms are used to stabilize replication and allow successful restart and completion of S phase at a later time.
References and Recommended Reading
Alcasabas, A. A. et al. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nature Cell Biology 3, 958–965 (2001).
Bando, M. et al. Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. Journal of Biological Chemistry 284, 34355–34365 (2009).
Bartkova, J. et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 (2005).
Bernstein, K. A. et al. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO Journal 28, 915–925 (2009).
Bianchi, V., Pontis, E., & Reichard, P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. Journal of Biological Chemistry 261, 16037–16042 (1986).
Boddy, M. N. et al. Replication checkpoint enforced by kinases Cds1 and Chk1. Science 280, 909–912 (1998).
Branzei, D. & Foiani, M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amsterdam) 6, 994–1003 (2007).
Branzei, D. & Foiani, M. The Rad53 signal transduction pathway: Replication fork stabilization, DNA repair, and adaptation. Experimental Cell Research 312, 2654–2659 (2006).
Brondello, J. M. et al. Basis for the checkpoint signal specificity that regulates Chk1 and Cds1 protein kinases. Molecular and Cellular Biology 19, 4262–4269 (1999).
Bryant, H. E. et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO Journal 28, 2601–2615 (2009).
Calzada, A. et al. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes & Development 19, 1905–1919 (2005).
Dalgaard, J. Z. & Klar, A. J. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102, 745–751 (2000).
Errico, A. & Costanzo, V. Differences in the DNA replication of unicellular eukaryotes and metazoans: Known unknowns. EMBO Reports 11, 270–278 (2010).
Errico, A., Costanzo, V., & Hunt, T. Tipin is required for stalled replication forks to resume DNA replication after removal of aphidicolin in Xenopus egg extracts. PNAS 104, 14929-14934 (2007).
Errico, A. et al. Tipin/Tim1/And1 protein complex promotes Pol alpha chromatin binding and sister chromatid cohesion. EMBO Journal 28, 3681–3692 (2009).
Feng, W. et al. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nature Cell Biology 8, 148–155 (2006).
Froget, B. et al. Cleavage of stalled forks by fission yeast Mus81/Eme1 in absence of DNA replication checkpoint. Molecular Biology of the Cell 19, 445–456 (2008).
Gambus, A. et al. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nature Cell Biology 8, 358–366 (2006).
Gambus, A. et al. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO Journal 28, 2992–3004 (2009).
Gotter, A. L., Suppa, C., & Emanuel, B. S. Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. Journal of Molecular Biology 366, 36–52 (2007).
Hanna, J. S. et al. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Molecular and Cellular Biology 21, 3144–3158 (2001).
Im, J. S. et al. Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. PNAS 106, 15628–15632 (2009).
Kai, M. et al. Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes & Development 19, 919–932 (2005).
Katou, Y. et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424, 1078–1083 (2003).
Kemp, M. G. et al. Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. Journal of Biological Chemistry 285, 16562–16571 (2010).
Koc, A. et al. Hydroxyurea arrests DNA replication by a mechanism that preserves basal dNTP pools. Journal of Biological Chemistry 279, 223–230 (2004).
Komata, M. et al. The direct binding of Mrc1, a checkpoint mediator, to Mcm6, a replication helicase, is essential for the replication checkpoint against methyl methanesulfonate-induced stress. Molecular and Cellular Biology 29, 5008–5019 (2009).
Koren, A., Soifer, I., & Barkai, N. MRC1-dependent scaling of the budding yeast DNA replication timing program. Genome Research 20, 781–790 (2010).
Krings, G. & Bastia, D. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. PNAS 101, 14085–14090 (2004).
Kumagai, A., Kim, S. M., & Dunphy, W. G. Claspin and the activated form of ATR-ATRIP collaborate in the activation of Chk1. Journal of Biological Chemistry 279, 49599–49608 (2004).
Kurose, A. et al. Effects of hydroxyurea and aphidicolin on phosphorylation of ataxia telangiectasia mutated on Ser 1981 and histone H2AX on Ser 139 in relation to cell cycle phase and induction of apoptosis. Cytometry Part A 69, 212–221 (2006).
Labib, K. Making connections at DNA replication forks: Mrc1 takes the lead. Molecular Cell 32, 166–168 (2008).
Labib, K. & Hodgson, B. Replication fork barriers: Pausing for a break or stalling for time? EMBO Reports 8, 346–353 (2007).
Langston, L. D., Indiani, C., & O'Donnell, M. Whither the replisome: Emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle 8, 2686–2691 (2009).
Lee, J. et al. Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Molecular Biology of the Cell 16, 5269–5282 (2005).
Leman, A. R. et al. Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. Journal of Cell Science 123, 660–670 (2010).
Lopes, M. et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412, 557–561 (2001).
Lou, H. et al. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Molecular Cell 32, 106–117 (2008).
Lucca, C. et al. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 23, 1206–1213 (2004).
Mamnun, Y. M., Katayama, S., & Toda, T. Fission yeast Mcl1 interacts with SCF(Pof3) and is required for centromere formation. Biochemical and Biophysical Research Communications 350, 125–130 (2006).
Mao, N., Kojic, M., & Holloman, W. K. Role of Blm and collaborating factors in recombination and survival following replication stress in Ustilago maydis. DNA Repair (Amsterdam) 8, 752–759 (2009).
Matsumoto, M., Rey, D. A., & Cory, J. G. Effects of cytosine arabinoside and hydroxyurea on the synthesis of deoxyribonucleotides and DNA replication in L1210 cells. Advances in Enzyme Regulation 30, 47–59 (1990).
McFarlane, R. J., Mian, S., & Dalgaard, J. Z. The many facets of the Tim-Tipin protein families' roles in chromosome biology. Cell Cycle 9, 700–705 (2010).
Meister, P. et al. Temporal separation of replication and recombination requires the intra-S checkpoint. Journal of Cell Biology 168, 537–544 (2005)
Miles, J. & Formosa, T. Evidence that POB1, a Saccharomyces cerevisiae protein that binds to DNA polymerase alpha, acts in DNA metabolism in vivo. Molecular and Cellular Biology 12, 5724–5735 (1992).
Mirkin, E. V. & Mirkin, S. M. Replication fork stalling at natural impediments. Microbiology and Molecular Biology Reviews 71, 13–35 (2007).
Mulder, K. W., Winkler, G. S., & Timmers, H. T. DNA damage and replication stress induced transcription of RNR genes is dependent on the Ccr4-Not complex. Nucleic Acids Research 33, 6384–6392 (2005).
Naylor, M. L. et al. Mrc1 phosphorylation in response to DNA replication stress is required for Mec1 accumulation at the stalled fork. PNAS 106, 12765–12770 (2009).
Nedelcheva, M. N. et al. Uncoupling of unwinding from DNA synthesis implies regulation of MCM helicase by Tof1/Mrc1/Csm3 checkpoint complex. Journal of Molecular Biology 347, 509–521 (2005).
Noguchi, C. & Noguchi, E. Sap1 promotes the association of the replication fork protection complex with chromatin and is involved in the replication checkpoint in Schizosaccharomyces pombe. Genetics 175, 553–566 (2007).
Noguchi, E. et al. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Molecular and Cellular Biology 24, 8342–8355 (2004).
Noguchi, E. et al. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Molecular and Cellular Biology 23, 7861–7874 (2003).
Osborn, A. J. & Elledge, S. J. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes & Development 17, 1755–1767 (2003).
Osborn, A. J., Elledge, S. J., & Zou, L. Checking on the fork: The DNA-replication stress-response pathway. Trends in Cell Biology 12, 509–516 (2002).
Petermann, E. et al. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Molecular Cell 37, 492–502 (2010).
Razidlo, D. F. & Lahue, R. S. Mrc1, Tof1 and Csm3 inhibit CAG. CTG repeat instability by at least two mechanisms. DNA Repair (Amsterdam) 7, 633–640 (2008).
Rothstein, R., Michel, B., & Gangloff, S. Replication fork pausing and recombination or "gimme a break." Genes & Development 14, 1–10 (2000).
Schleker, T., Nagai, S., & Gasser, S. M. Posttranslational modifications of repair factors and histones in the cellular response to stalled replication forks. DNA Repair (Amsterdam) 8, 1089–1100 (2009).
Smith, K. D., Fu, M. A. & Brown, E. J. Tim-Tipin dysfunction creates an indispensible reliance on the ATR-Chk1 pathway for continued DNA synthesis. Journal of Cell Biology 187, 15–23 (2009).
Szyjka, S. J., Viggiani, C. J., & Aparicio, O. M. Mrc1 is required for normal progression of replication forks throughout chromatin in S. cerevisiae. Molecular Cell 19, 691–697 (2005).
Tanaka, K. & Russell, P. Cds1 phosphorylation by Rad3-Rad26 kinase is mediated by forkhead-associated domain interaction with Mrc1. Journal of Biological Chemistry 279, 32079–32086 (2004).
Tanaka, K. & Russell, P. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nature Cell Biology 3, 966–972 (2001).
Tanaka, H., Katou, Y., et al. Ctf4 coordinates the progression of helicase and DNA polymerase alpha. Genes to Cells 14, 807–820 (2009).
Tanaka, H., Kubota, Y., et al. Replisome progression complex links DNA replication to sister chromatid cohesion in Xenopus egg extracts. Genes to Cells 14, 949–963 (2009).
Torres-Rosell, J., De Piccoli, G., & Aragon, L. Can eukaryotic cells monitor the presence of unreplicated DNA? Cell Division 2, 19 (2007).
Tourriere, H. & Pasero, P. Maintenance of fork integrity at damaged DNA and natural pause sites. DNA Repair (Amsterdam) 6, 900–913 (2007).
Tourriere, H. et al. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Molecular Cell 19, 699–706 (2005).
Unsal-Kacmaz, K. et al. The human Tim/Tipin complex coordinates an intra-S checkpoint response to UV that slows replication fork displacement. Molecular and Cellular Biology 27, 3131–3142 (2007).
Urtishak, K. A. et al. Timeless maintains genomic stability and suppresses sister chromatid exchange during unperturbed DNA replication. Journal of Biological Chemistry 284, 8777–8785 (2009).
Williams, D. R. & McIntosh, J. R. mcl1+, the Schizosaccharomyces pombe homologue of CTF4, is important for chromosome replication, cohesion, and segregation. Eukaryotic Cell 1, 758–773 (2002).
Wray, J. et al. Distinct RAD51 associations with RAD52 and BCCIP in response to DNA damage and replication stress. Cancer Research 68, 2699–2707 (2008).
Xu, Y. J., Davenport, M., & Kelly, T. J. Two-stage mechanism for activation of the DNA replication checkpoint kinase Cds1 in fission yeast. Genes & Development 20, 990–1003 (2006).
Yoshizawa-Sugata, N. & Masai, H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. Journal of Biological Chemistry 282, 2729–2740 (2007).
Yoshizawa-Sugata, N. & Masai, H. Roles of human AND-1 in chromosome transactions in S phase. Journal of Biological Chemistry 284, 20718–20728 (2009).
Zhao, H. et al. Replication checkpoint protein Mrc1 is regulated by Rad3 and Tel1 in fission yeast. Molecular and Cellular Biology 23, 8395–8403 (2003).
Zhu, W. et al. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes & Development 21, 2288–2299 (2007).


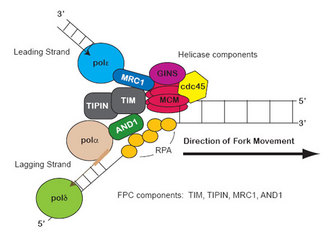
 Figure 1: Replication fork components
Figure 1: Replication fork components


























