« Prev Next »

In 1931, Dorothea Smith observed variable mitochondrial networks in developing rat liver cells (Smith 1931). Over subsequent years, many scientists described complex mitochondrial networks in numerous cell types (reviewed in Bereiter-Hahn & Voth 1994). A mitochondrial network consists of a complex, branched system of connected mitochondria (Figure 1). Some modern textbooks still depict mitochondria inaccurately as small, static ovoid organelles, perhaps because early cell cross sections appeared to show many distinct mitochondria, which we now know are often connected in a plane outside the one visualized. Mitochondrial fusion is the physical merging of the outer and then the inner mitochondrial membranes of two originally distinct mitochondria. Mitochondrial division is the breaking apart of one mitochondrial body into two. The combined effects of ongoing fusion and division give rise to mitochondrial networks.
Why Are Dynamic Mitochondrial Networks Evolutionarily Conserved?
Mitochondrial connectivity allows for even distribution of special mitochondrial components such as the lipid cardiolipin. Fusion of mitochondria also enables genetic complementation: two mitochondrial genomes with different defects inside the same organelle can each encode what the other lacks, generating all components for a functional mitochondrion. As hypothesized by Vladimir Skulachev (2001), a branched mitochondrial network may allow ATP generation in oxygen-poor regions of a cell through electrical transmission of the mitochondrial membrane potential across the network. In addition, mitochondrial division facilitates the transport of mitochondria to daughter cells during mitosis and meiosis.
Balance of Mitochondrial Fusion and Division
Mitochondrial fusion and division typically counterbalance each other. Levels of nutrients and other criteria can tip the balance toward either fusion or division. In some specialized cell types, like neurons and sperm, energy needs are high and mitochondria are trafficked long distances, dictating unique needs for the shape of the mitochondrial network. Researchers have identified many gene products that control mitochondrial fusion and division. Three central players are (1) mitofusins (outer mitochondrial membrane fusion), (2) OPA1/Mgm1 (inner mitochondrial membrane fusion), and (3) Drp1/Dnm1 (division of outer and inner mitochondrial membranes). All of these molecules are GTP-hydrolyzing proteins (GTPases) that belong to the dynamin superfamily. The different regulation of these proteins and their binding partners is a framework that scientists use to understand mitochondrial dynamics in different cells and contexts.
How Did We Come to Understand Mitochondrial Fusion?
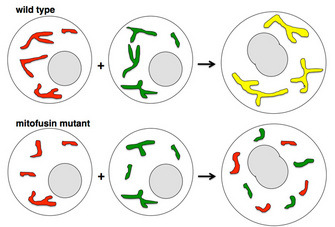
All membrane fusion events in the cell are tightly regulated by protein mediators. Otherwise, a hopeless jumble of structures would result from the random fusion of any two nearby membrane-enclosed organelles. The proteins that enable mitochondria to fuse are different from those that allow fusion between other structures, such as Golgi-derived vesicles. How do scientists identify these mitochondrial fusion mediators? They use a genetic approach and start with mutant strains of a model organism that have defects in mitochondrial fusion. For example, in the maturing sperm cells of the fruit fly Drosophila melanogaster, mitochondria undergo dramatic shaping, including aggregation, fusion, and elongation. Defects lead to sterility, making the fruit fly a good system for genetic study of mitochondrial shaping, since male sterile strains can be easily identified from thousands of strains with different random mutations. Karen Hales and Margaret Fuller at Stanford University used this model system to discover the first known protein mediator of mitochondrial fusion. All the mitochondria in a Drosophila spermatid normally fuse into two large, intertwined mitochondrial derivatives, which then disentangle and elongate as the sperm tail grows. Male flies homozygous for mutations in the fzo gene are sterile because mitochondria fail to fuse and mature sperm are nonmotile. Hales and Fuller used genetic mapping and cloning techniques with this strain to identify the fzo gene, which encodes the founding member of the conserved mitofusin GTPase family (Hales & Fuller 1997).
A Lack of Functional Mitofusin Can Reveal Its Essential Role
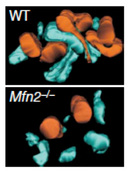
As it turns out, mitofusin defects can also occur in humans. The human neurodegenerative disorder Charcot-Marie-Tooth disease type 2A, characterized by progressive sensory and motor losses in the extremities, results from mutations in human mitofusin Mfn2 (Zuchner et al. 2004). Scientists have generated mouse strains carrying mutations that directly mimic mutations in human patients, and the mouse and human symptoms are similar (Detmer et al. 2008). Even with a mouse model, however, scientists face significant challenges in devising mitochondrial treatments (and delivering them to the relevant neurons) in Charcot-Marie-Tooth type 2A patients.
How Do Mitofusins Work?
What Role Do Mitofusins Play in Mitochondria-ER Tethering and Mitochondrial Transport?
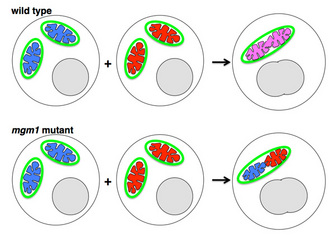
Fusion of the Inner Mitochondrial Membrane: Role of OPA1/Mgm1
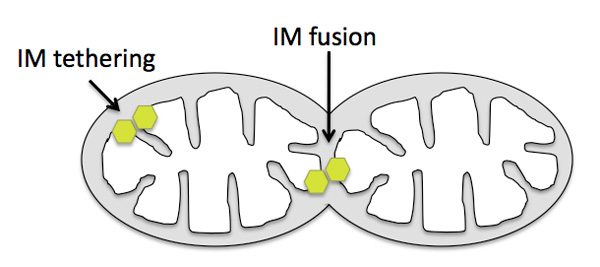
If OPA1/Mgm1 promotes fusion of inner mitochondrial membranes, then why don't the cristae (infoldings of inner membrane) fuse together? In fact, OPA1/Mgm1 does indeed tether (but not fuse) cristae and helps maintain their folded structure (Figure 7). Several research groups showed by immuno-electron microscopy that the OPA1/Mgm1 protein is present on cristae and that in mutant cells lacking the protein, the cristae membranes are not held together properly (Griparic et al. 2004; Olichon et al. 2003; Wong et al. 2000). As with mitofusins, we do not yet know what controls OPA1/Mgm1-mediated tethering versus fusion. Nunnari and colleagues speculate that OPA1/Mgm1-mediated inner membrane fusion may occur only after a signal from active mitofusins on the outer membrane (Hoppins et al. 2007). OPA1/Mgm1 is thus a dual-function protein important both for inner mitochondrial membrane fusion and for maintaining the shape of cristae. Defects in either or both processes underlie the progressive blindness disorder dominant optic atrophy, which is caused by OPA1 mutations in humans (Alexander et al. 2000; Delettre et al. 2000).
Division of Mitochondria: Role of Drp1/Dnm1
Mitochondria are not always fusing. They have an equal, balanced activity of division (fission) within most cells. Why do mitochondria divide? Mitochondrial division is important for the remodeling and rearrangement of mitochondrial networks, as well as for enabling mitochondrial segregation during cell division. William Bleazard and colleagues in Janet Shaw's lab, and Hiromi Sesaki in Rob Jensen's lab at Johns Hopkins University, defined a role in yeast mitochondrial division for a conserved, large dynamin-related GTPase called Dnm1 (Drp1 in mammals). They discovered that, in cells with mutations in the dnm1 gene, mitochondria form large nets that result from failed mitochondrial division and therefore unopposed mitochondrial fusion; the system of fission and fusion is thrown out of balance (Figure 3; Bleazard et al. 1999; Sesaki & Jensen 1999). Other researchers confirmed a mitochondrial division role for homologs of this protein in different organisms (reviewed in Hoppins et al. 2007; Liesa et al. 2009).
Curiously, researchers who made yeast strains with fluorescently tagged Dnm1 found that not all the protein in the cell associates with mitochondria (Otsuga et al. 1998; Sesaki & Jensen 1999). Some of it was observed in the cytoplasm, while some was in clumps on the mitochondrial surface at mitochondrial division sites. Perhaps this is not surprising, since the protein does not include a transmembrane domain and is therefore not expected to be permanently anchored in a membrane on its own. Several research groups discovered that a different gene product, called Fis1, which does have a transmembrane domain, sits in the outer mitochondrial membrane and is crucial for recruiting Drp1/Dnm1 to mitochondrial fission sites. In yeast cells lacking Fis1, the Drp1/Dnm1 protein was present but could not associate with mitochondria, and mitochondria formed a net structure representing failed mitochondrial division (James et al. 2003; Mozdy et al. 2000; Yoon et al. 2003).
Regulation of Mitochondrial Fusion and Division
Up-and-down regulation of mitofusins, OPA1/Mgm1, and Drp1/Dnm1 cumulatively dictate the balance between mitochondrial fusion and division in different cell contexts. For example, mitochondria break into smaller pieces early in programmed cell death (apoptosis). This event is triggered by up-regulation of Drp1/Dnm1 and down-regulation of mitofusins. Later in apoptosis, altered OPA1/Mgm1 activity in the inner mitochondrial membrane causes a change in cristae structure, release of cytochrome c, and subsequent activation of the destructive caspase enzymes (reviewed in Suen et al. 2008), indicating that inner mitochondrial membrane structure is intertwined with regulatory pathways influencing cell life and death.
Regulation of these three mitochondrial shaping proteins occurs in different ways in various cell contexts and at many levels, including protein stability, protein cleavage, protein conformation via covalent modification, and protein localization via association with binding partners. The additional protein participants are too numerous to describe here. In brief, degradation of mitofusins by the ubiquitin-proteasome system determines their steady-state levels and thus influences mitochondrial fusion (Cohen et al. 2008). OPA1/Mgm1 exists as several different-sized isoforms with overlapping functions; proteases that generate these isoforms regulate OPA1/Mgm1 activity (Griparic et al. 2007; Ishihara et al. 2006). The mitochondrial fission mediator Drp1/Dnm1 is regulated by posttranslational modifications such as phosphorylation and the addition of the ubiquitin-like protein SUMO (reviewed in Santel & Frank 2008), as well as by association with binding partners on the mitochondrial surface.
Possible Misregulation of Mitochondrial Fusion in Parkinson's Disease
Finally, the common motor neurodegenerative disorder Parkinson's disease appears to be related to the control of mitochondrial dynamics, at least in some patients. Normally, mitochondrial fusion enhances mitochondrial integrity by allowing component sharing across the tubular network. However, fusion of highly damaged mitochondria to the network could be detrimental, since hobbled mitochondria generate reactive oxygen species that cause a chain reaction of damage. Cells have mechanisms to segregate and degrade badly damaged mitochondria. Two research groups (Angela Poole and colleagues in Leo Pallanck's laboratory at the University of Washington, and Elena Ziviani and Ran Tao in Alexander Whitworth's laboratory at the University of Sheffield) showed that mitochondrial quality control involves mitofusin regulation by the Parkinson's-associated gene products PINK1 and Parkin (Poole et al. 2010; Ziviani et al. 2010). In normal cells, PINK1 senses a membrane potential change on damaged mitochondria and recruits Parkin, which in turn ubiquitinates mitofusins. These damaged, ubiquitin-marked mitochondria are then targeted for degradation. A current hypothesis, not yet proven, is that alteration of mitofusins may also prevent damaged mitochondria from fusing to the healthy network. In Parkinson's patients with faulty PINK1 or Parkin, damaged mitochondria are not degraded and remain integrated in the network, leading to cell damage and neurodegeneration.
Summary
Mitochondrial fusion and division are mediated by several large GTPases whose combined effects lead to the dynamic mitochondrial networks seen in many cell types. Mitofusins control outer mitochondrial membrane fusion, and OPA1/Mgm1 mediates inner mitochondrial membrane fusion. Drp1/Dnm1 is a central player in mitochondrial division. Many additional protein participants modulate the activities of these proteins in various contexts. Several human genetic neurodegenerative diseases are associated with improper regulation of mitochondrial fusion or division. Scientists are currently determining how the molecular shape changes of these GTPases control the precise three-dimensional arrangement of phospholipids during fusion or division. Scientists are also searching for additional ways in which these fusion and division mediators are regulated in different tissues, since each mechanism of regulation represents a possible therapeutic target for patients with related disorders.
References and Recommended Reading
Alexander, C. et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nature Genetics 26, 211–215 (2000).
Baloh, R. H. et al. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. Journal of Neuroscience 27, 422–430 (2007) doi:10.1523/JNEUROSCI.4798-06.2007.
Bereiter-Hahn, J. & Voth, M. Dynamics of mitochondria in living cells: Shape changes, dislocations, fusion, and fission of mitochondria. Microscopy Research and Technique 27, 198–219 (1994).
Bleazard, W. et al. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nature Cell Biology 1, 298–304 (1999).
Cerveny, K. L. et al. Regulation of mitochondrial fusion and division. Trends in Cell Biology 17, 563–569 (2007) doi:10.1016/j.tcb.2007.08.006.
Chen, H. & Chan, D. C. Mitochondrial dynamics — fusion, fission, movement, and mitophagy — in neurodegenerative diseases. Human Molecular Genetics 18, R169–R176 (2009) doi:10.1093/hmg/ddp326.
Chen, H., McCaffery, J. M. & Chan, D. C. Mitochondrial fusion protects against neurodegeneration in the cerebellum. Cell 130, 548–562 (2007) doi:10.1016/j.cell.2007.06.026.
Chen, H. et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell 141, 280–289 (2010) doi:10.1016/j.cell.2010.02.026.
Chen, H. et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. Journal of Cell Biology 160, 189–200 (2003).
Cohen, M. M. et al. Ubiquitin-proteasome-dependent degradation of a mitofusin, a critical regulator of mitochondrial fusion. Molecular Biology of the Cell 19, 2457–2464 (2008) doi:10.1091/mbc.E08-02-0227.
de Brito, O. M. & Scorrano, L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610 (2008) 10.1038/nature07534.
Delettre, C. et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nature Genetics 26, 207–210 (2000) doi:10.1038/79936.
Detmer, S. A. & Chan, D. C. Functions and dysfunctions of mitochondrial dynamics. Nature Reviews Molecular Cell Biology 8, 870–879 (2007) doi:10.1038/nrm2275.
Detmer, S. A. et al. Hindlimb gait defects due to motor axon loss and reduced distal muscles in a transgenic mouse model of Charcot-Marie-Tooth type 2A. Human Molecular Genetics 17, 367–375 (2008) doi:10.1093/hmg/ddm314.
Griparic, L., Kanazawa, T. & van der Bliek, A. M. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. Journal of Cell Biology 178, 757–764 (2007) doi:10.1083/jcb.200704112.
Griparic, L. et al. Loss of the intermembrane space protein Mgm1/OPA1 induces swelling and localized constrictions along the lengths of mitochondria. Journal of Biological Chemistry 279, 18792–18798 (2004).
Hales, K. G. & Fuller, M. T. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90, 121–129 (1997).
Hermann, G. J. et al. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. Journal of Cell Biology 143, 359–373 (1998).
Hoppins, S., Lackner, L. & Nunnari, J. The machines that divide and fuse mitochondria. Annual Review of Biochemistry 76, 751–780 (2007) doi:10.1146/annurev.biochem.76.071905.090048.
Ishihara, N. et al. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO Journal 25, 2966–2977 (2006) doi:10.1038/sj.emboj.7601184.
James, D. I. et al. hFis1, a novel component of the mammalian mitochondrial fission machinery. Journal of Biological Chemistry 278, 36373–36379 (2003).
Koshiba, T. et al. Structural basis of mitochondrial tethering by mitofusin complexes. Science 305, 858–862 (2004) doi:10.1126/science.1099793.
Liesa, M., Palacin, M. & Zorzano, A. Mitochondrial dynamics in mammalian health and disease. Physiological Reviews 89, 799–845 (2009) doi:10.1152/physrev.00030.2008.
Mears, J. A. & Hinshaw, J. E. Visualization of dynamins. Methods in Cell Biology 88, 237–256 (2008) doi:10.1016/S0091-679X(08)00413-5.
Meeusen, S. et al. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell 127, 383–395 (2006) doi:10.1016/j.cell.2006.09.021.
Misko, A. et al. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. Journal of Neuroscience 30, 4232–4240 (2010) doi:10.1523/JNEUROSCI.6248-09.2010.
Mozdy, A. D., McCaffery, J. M. & Shaw, J. M. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. Journal of Cell Biology 151, 367–380 (2000).
Nunnari, J. et al. Mitochondrial transmission during mating in Saccharomyces cerevisiae is determined by mitochondrial fusion and fission and the intramitochondrial segregation of mitochondrial DNA. Molecular Biology of the Cell 8, 1233–1242 (1997).
Olichon, A. et al. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. Journal of Biological Chemistry 278, 7743–7746 (2003).
Otsuga, D. et al. The dynamin-related GTPase, Dnm1p, controls mitochondrial morphology in yeast. Journal of Cell Biology 143, 333–349 (1998).
Poole, A. C. et al. The mitochondrial fusion-promoting factor mitofusin is a substrate of the PINK1/parkin pathway. PLoS One 5, e10054 (2010) doi:10.1371/journal.pone.0010054.
Santel, A. & Frank, S. Shaping mitochondria: The complex posttranslational regulation of the mitochondrial fission protein DRP1. IUBMB Life 60, 448–455 (2008) .doi:10.1002/iub.71.
Sesaki, H. & Jensen, R. E. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. Journal of Cell Biology 147, 699–706 (1999).
Shaw, J. M. & Winge, D. R. Shaping the mitochondrion: Mitochondrial biogenesis, dynamics and dysfunction. Conference on Mitochondrial Assembly and Dynamics in Health and Disease. EMBO Reports 10, 1301–1305 (2009) doi:10.1038/embor.2009.247.
Skulachev, V. P. Mitochondrial filaments and clusters as intracellular power-transmitting cables. Trends in Biochemical Sciences 26, 23–29 (2001).
Smith, D. M. The ontogenic history of the mitochondria of the hepatic cell of the white rat. Journal of Morphology and Physiology 52, 485–512 (1931).
Suen, D. F., Norris, K. L. & Youle, R. J. Mitochondrial dynamics and apoptosis. Genes & Development 22, 1577–1590 (2008) doi:10.1101/gad.1658508.
Westermann, B. Merging mitochondria matters: Cellular role and molecular machinery of mitochondrial fusion. EMBO Reports 3, 527–531 (2002).
Westermann, B. Mitochondrial dynamics in model organisms: What yeasts, worms and flies have taught us about fusion and fission of mitochondria. Seminars in Cell & Developmental Biology 21, 542–549 (2010) doi:10.1016/j.semcdb.2009.12.003.
Westermann, B. Molecular machinery of mitochondrial fusion and fission. Journal of Biological Chemistry 283, 13501–13505 (2008) doi:10.1074/jbc.R800011200.
Wong, E. D. et al. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. Journal of Cell Biology 151, 341–352 (2000).
Yoon, Y. et al. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Molecular and Cellular Biology 23, 5409–5420 (2003).
Ziviani, E., Tao, R. N. & Whitworth, A. J. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. PNAS 107, 5018–5023 (2010) doi:10.1073/pnas.0913485107.
Zuchner, S. et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nature Genetics 36, 449–451 (2004).



 Figure 1: Mitochondrial networks
Figure 1: Mitochondrial networks