« Prev Next »
One important instance of recombination in diploid eukaryotic organisms is the exchange of genetic information between newly duplicated chromosomes during the process of meiosis. In this instance, the outcome of recombination is to ensure that each gamete includes both maternally and paternally derived genetic information, such that the resulting offspring will inherit genes from all four of its grandparents, thereby acquiring a maximum amount of genetic diversity. Recombination is also used in DNA repair (particularly in the repair of double-stranded breaks), as well as during DNA replication to assist in filling gaps and preventing stalling of the replication fork. In these cases, a sister chromatid serves as the donor of missing material via recombination followed by DNA synthesis.
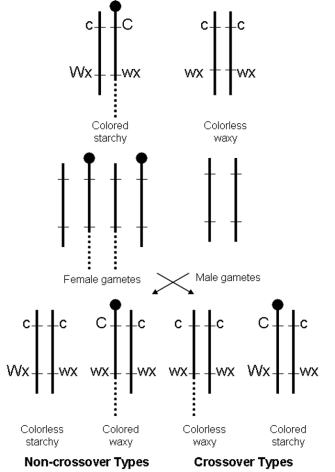
The role of recombination during the inheritance of chromosomes was first demonstrated through experiments with maize. Specifically, in 1931, Barbara McClintock and Harriet Creighton obtained evidence for recombination by physically tracking an unusual knob structure within certain maize chromosomes through multiple genetic crosses. Using a strain of maize in which one member of a chromosome pair exhibited the knob but its homologue did not, the scientists were able to show that some alleles were physically linked to the knobbed chromosome, while other alleles were tied to the normal chromosome. McClintock and Creighton then followed these alleles through meiosis, showing that alleles for specific phenotypic traits were physically exchanged between chromosomes. Evidence for this finding came from the fact that alleles first introduced into the cross on a knobbed chromosome later appeared in offspring without the knob; similarly, alleles initially introduced on a knobless chromosome subsequently appeared in progeny with the knob (Figure 1).
Recombination also occurs in prokaryotic cells, and it has been especially well characterized in E. coli. Although bacteria do not undergo meiosis, they do engage in a type of sexual reproduction called conjugation, during which genetic material is transferred from one bacterium to another and may be recombined in the recipient cell. As in eukaryotes, recombination also plays important roles in DNA repair and replication in prokaryotic organisms.
Models of Recombination
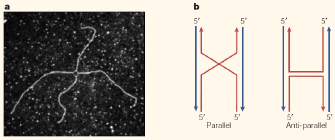
Although common, genetic recombination is a highly complex process. It involves the alignment of two homologous DNA strands (the requirement for homology suggests that this occurs through complementary base-pairing, but this has not been definitively shown), precise breakage of each strand, exchange between the strands, and sealing of the resulting recombined molecules. This process occurs with a high degree of accuracy at high frequency in both eukaryotic and prokaryotic cells.
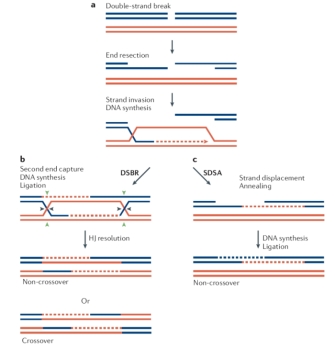
The basic steps of recombination can occur in two pathways, according to whether the initial break is single or double stranded. In the single-stranded model, following the alignment of homologous chromosomes, a break is introduced into one DNA strand on each chromosome, leaving two free ends. Each end then crosses over and invades the other chromosome, forming a structure called a Holliday junction (Figure 2). The next step, called branch migration, takes place as the junction travels down the DNA. The junction is then resolved either horizontally, which produces no recombination, or vertically, which results in an exchange of DNA.
In the alternate pathway initiated by double-stranded breaks, the ends at the breakpoints are converted into single strands by the addition of 3' tails. These ends can then perform strand invasion, producing two Holliday junctions. From that point forward, resolution proceeds as in the single-stranded model (Figure 3). (Note that a third model of recombination, synthesis-dependent strand annealing [SDSA], has also been proposed to account for the lack of crossover typical of recombination in mitotic cells and observed in some meiotic cells to a lesser degree.)
Recombination Enzymes
No matter which pathway is used, a number of enzymes are required to complete the steps of recombination. The genes that code for these enzymes were first identified in E. coli by the isolation of mutant cells that were deficient in recombination. This research revealed that the recA gene encodes a protein necessary for strand invasion. Meanwhile, the recB, recC, and recD genes code for three polypeptides that join together to form a protein complex known as RecBCD; this complex has the capacity to unwind double-stranded DNA and cleave strands. Two other genes, ruvA and ruvB, encode enzymes that catalyze branch migration, while Holliday structures are resolved by the protein resolvase, which is product of the ruvC gene. Several enzymes involved in DNA replication, such as ligase and DNA polymerase, also contribute to recombination (Clark, 1973).
In eukaryotes, recombination has been perhaps most thoroughly studied in the budding yeast Saccharomyces cerevisiae. Many of the enzymes identified in this yeast have also been found in other organisms, including mammalian cells. Such studies reveal that the Rad genes (named for the fact that their activity was found to be sensitive to radiation) play a key role in eukaryotic recombination. In particular, the Rad51 gene, which is homologous to recA, encodes a protein (called Rad51) that has recombinase activity. This gene is highly conserved, but the accessory proteins that assist Rad51 appear to vary among organisms. For example, the Rad52 protein is found in both yeast and humans, but it is missing in Drosophila melanogaster and C. elegans.
In eukaryotic cells, single-stranded DNA (ssDNA) becomes rapidly coated with the protein RPA (replication protein A). RPA has a higher affinity for ssDNA than Rad51, and it therefore can inhibit recombination by blocking Rad51's access to the single strand needed for invasion. In yeast, however, binding of Rad51 to ssDNA is enhanced by the proteins Rad52 and the complex Rad55-Rad57. Once access has been gained, Rad51 polymerizes on the DNA strand to form what is called a presynaptic filament, which is a right-handed helical filament containing six Rad51 molecules and 18 nucleotides per helical repeat. The search for DNA homology and formation of the junction between homologous regions is then carried out within the catalytic center of the filament.
In addition to proteins that assist Rad51 activity, there are also some proteins that inhibit it. In yeast, for instance, the helicase Srs2 dismantles the Rad51-ssDNA complex, while the proteins Sgs1 and BLM inhibit the complex. It is thought that these proteins play a role in preventing recombination during DNA replication when it is not needed.
In humans, the tumor suppressor genes BRCA1 and BRCA2 also play a role in regulating recombination. Individuals who are heterozygous for BRCA2 are subject to increased risk for breast and ovarian cancer; loss of both alleles causes Fanconi's anemia, a genetic disease characterized by predisposition to cancer, among other defects. BRCA2 appears to promote Rad51 binding to ssDNA (Li & Heyer, 2008; Modesti & Kanaar, 2001).
How Are Homologous Sequences Brought Together?
As previously described, the enzymes and mechanisms that carry out the process of homologous recombination are fairly well delineated. Not so well understood is the important question of how homologous sequences come to be in proximity so that recombination can proceed. In their 2008 review, Barzel and Kupiec describe two alternate hypotheses, one of which they call the null model. This model proposes that homologues find one another through a passive process of diffusion, in which the DNA sequence at the broken end of a strand is sequentially compared to all of the other potential end sequences in the genome. In order for diffusion to account for the rapid repair of double-stranded breaks observed in yeast, however, Barzel and Kupiec calculate that each homology search would have to proceed at a speed 40 times faster than the rate at which DNA polymerase adds a single nucleotide to a replicating DNA chain, which seems unlikely (Barzel & Kupiec, 2008).
An alternate hypothesis proposes that homologous chromosomes reside in pairs constitutively. Acting against this hypothesis is the finding that in induced recombination experiments, the broken ends of strands recombine with what are called ectopic homologues (areas of fortuitous sequence identity) as frequently as they recombine with their true homologous chromosomes. Furthermore, although homologous pairing has been observed in somatic cells of some organisms (e.g., Drosophila, Neurospora), it is not widely seen in the cells of other organisms, including mammals. As Barzel and Kupiec (2008) point out, the absence of general homologous pairing does not necessarily mean random assortment. Instead, discrete sections of chromosomes may be required for homology. The use of subdomains for homology searches would reduce the time it takes to find a homologous partner. Despite such theories, the exact mechanism responsible for locating and lining up homologous segments remains to be determined.
References and Recommended Reading
Barzel, A., & Kupiec, M. Finding a match: How do homologous sequences get together for recombination? Nature Reviews Genetics 9, 27–37 (2008) doi:10.1038/nrg2224 (link to article)
Clark, A. J. 1973. Recombination deficient mutants of E. coli and other bacteria. Annual Review of Genetics 7, 67–86 (1973) doi:10.1146/annurev.ge.07.120173.000435
Li, X., & Heyer, W. D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Research 18, 99–113 (2008)
Liu, Y., & West, S. C. Happy Hollidays: Fortieth anniversary of the Holliday junction. Nature Reviews Molecular Cell Biology 5, 937–944 (2004) doi:10.1038/nrm1502 (link to article)
Modesti, M., & Kanaar, R. Homologous recombination: From model organism to human disease. Genome Biology 2, 1014.1–1014.5 (2001)
Sung, P., & Klein, H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nature Reviews Molecular Cell Biology 7, 739–750 (2006) doi:10.1038/nrm2008 (link to article)



 Figure 1: McClintock and Creighton's work in maize shows physical evidence of recombination.
Figure 1: McClintock and Creighton's work in maize shows physical evidence of recombination.