« Prev Next »
All of the cells within a complex multicellular organism such as a human being contain the same DNA; however, the body of such an organism is clearly composed of many different types of cells. What, then, makes a liver cell different from a skin or muscle cell? The answer lies in the way each cell deploys its genome. In other words, the particular combination of genes that are turned on (expressed) or turned off (repressed) dictates cellular morphology (shape) and function. This process of gene expression is regulated by cues from both within and outside cells, and the interplay between these cues and the genome affects essentially all processes that occur during embryonic development and adult life.
Do All Cells Really Contain the Same DNA?

Several lines of evidence support the proposal that all of the cells within a multicellular organism contain the same genome. For instance, although you started as a single cell with a half-genome from each parent, that single cell quickly divided and new cells began to differentiate, or become different from each other. While this process of differentiation established a wide variety of cell types (e.g., skin, liver, muscle, etc.), it was not accompanied by any permanent loss of genetic material. This is demonstrated by the fact that fully differentiated cell types are still capable, within the right environment, of giving rise to an entire new animal. This capability was first shown by way of an experiment in which the nucleus of an adult frog skin cell was transplanted into an enucleated donor embryo, eventually leading to the development of a cloned adult frog (Gurdon et al., 1975). Later, the intact complete genome of a differentiated cell was used in the cloning of the famous sheep Dolly (Figure 1), showing that in mammals, genes are not lost during development, so they must therefore be regulated (Wilmut et al., 1997).
Today, researchers understand that the specialized, differentiated cell types of the adult body contain a genome as complete as any embryo's. This fascinating demonstration has led to the proposal that changes in gene expression, rather than losses of genetic material, play a key role in guiding and maintaining cell differentiation.
Cell-Extrinsic Regulation of Gene Expression
Gene expression is regulated by factors both extrinsic and intrinsic to the cell. Cell-extrinsic factors that regulate expression include environmental cues, such as small molecules, secreted proteins, temperature, and oxygen. These cues can originate from other cells within the organism, or they can come from the organism's environment. Within the organism, cells communicate with each other by sending and receiving secreted proteins, also known as growth factors, morphogens, cytokines, or signaling molecules. Receipt of these signaling molecules triggers intercellular signaling cascades that ultimately cause semipermanent changes in transcription or expression of genes. Such changes in gene expression can include turning genes completely on or off, or just slightly tweaking the level of transcript produced. This process is thought to regulate a vast number of cell behaviors, including cell fate decisions during embryogenesis, cell function, and chemotaxis.
In addition, gene expression changes can lead to changes in an entire organism, such as molting in insects. In Drosophila, for example, the molting process is regulated by levels of a hormone called ecdysone. This hormone acts as a signal, triggering a cascade of events and leading to changes in gene expression. Not surprisingly, the genes that are expressed in response to ecdysone are also the genes that are involved in the molting process (White et al., 1997). Thus, ecdysone acts on the organism level as a cell-extrinsic factor to bring about physiologically meaningful changes in gene expression.
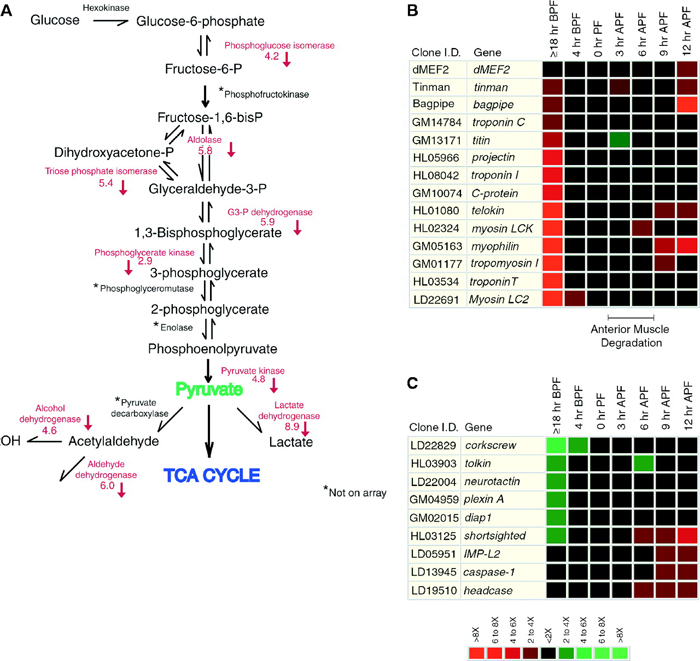
What is also interesting is that scientists can learn more about a physiological process like metamorphosis by studying how gene expression patterns change over time. For example, although researchers were aware that ecdysone results in a decrease of transcription from some loci, such as those involved in the glycolytic pathway, microarray data suggest that ecdysone-induced metamorphosis also downregulates genes involved with fatty acid oxidation, amino acid metabolism, oxidative phosphorylation, and other pathways. This suggests that there is a more global repression of metabolic activity during molting (Figure 2). Specifically, during metamorphosis, the larval muscle cells are degraded, and muscle-specific genes are downregulated (Figure 2B). Simultaneously, the development of the nervous system begins, and the genes involved in neuronal differentiation are induced (Figure 2C).
Cell-Intrinsic Regulation of Gene Expression
Although differentiation is not thought to occur by permanent loss of genetic material, DNA can be modified in a way that affects gene expression. For instance, DNA and its associated histone proteins (together known as chromatin) can be chemically modified by a cell's own machinery. Chromatin modification can affect gene expression by changing the accessibility of genes to transcription factors, in either a positive or a negative manner. Two major classes of such chemical modifications include DNA methylation and histone modification (methylation and/or acetylation). These changes are often described as epigenetic because they do not act to alter the primary DNA sequence but instead act at a level just above the DNA sequence. Although DNA methylation and histone modification are not genetic, cells have mechanisms to copy this epigenetic information during their division so that their daughter cells contain the same regulatory data.
Changes in chromatin modification play an important role in regulating gene expression during developmental cell-type specification as well. For example, chromatin-modifying proteins play an essential role in muscle cell differentiation via interactions with key muscle-promoting transcription factors MyoD and MEF. That is, these factors are thought to help recruit chromatin modifying factors, such as histone acetyltransferases and deacetylases. In so doing, MyoD and MEF alter access to their target sites upstream of muscle differentiation genes. For instance, MyoD binds histone acetyltransferases p300 and PCAF, and this activity is essential for muscle cell differentiation (Puri et al., 1997). This example provides evidence for a link among chromatin modifications, transcription factors, and, ultimately, cell-fate-specific changes in gene expression.
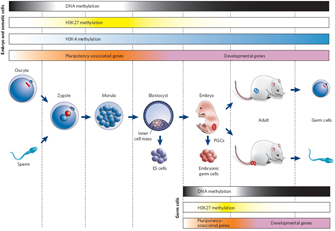
Chromatin modification can be stable over the life of an organism, thereby effectively permanently influencing gene expression. However, that is not to say that chromatin modification is irreversible. For instance, chromatin can become mismodified in certain cancers (Vucic et al., 2008), suggesting that, although important, the change is not permanent. Moreover, chromatin modifications are usually erased and reset during the production of gametes, such that the adult program of intrinsic cues is replaced with a program more suited to embryonic development (Figure 3; Reik, 2007).
In fact, embryonic cell types are known to contain a unique set of chromatin modifications that are different from those found in adult cell types (Bernstein et al., 2006; Meshorer et al., 2006). This has led to the tantalizing proposal that chromatin modification helps lock in changes in gene expression that are required during development. The permanent silencing of the genes involved only in embryogenesis could then drive the development of cells toward more mature cell types. By blocking accessibility of transcription machinery, for example, chromatin modification could prevent the need for continued repression through active binding of a repressive transcription factor. Alternatively, the genes required for an adult cell type might contain chromatin modifications (especially histone acetylation) that cause the DNA to become open and, therefore, more accessible to the transcription machinery.
Interestingly, embryonic cell types have been found to contain a signature chromatin modification in the regions that regulate the expression of genes involved in early embryonic development (Bernstein et al., 2006). Such regions were found to contain chromatin modifications with both silencing and promoting characteristics. The finding of these bivalent (two-directional) markers in association with genes important for embryonic development has led to the belief that embryonic cells exist in a special epigenetic state, wherein they can choose to remain embryonic (as in an embryonic stem cell) or to differentiate (as in normal development), and bivalent domains provide a means by which to quickly choose between the two options.
Together, these lines of evidence have led to an emerging hypothesis that cell-cell signaling and epigenetic changes converge to guide cell differentiation decisions both during development and beyond.
References and Recommended Reading
Bernstein, B. E., et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006)
Giles, J., et al. Dolly's death leaves researchers woolly on clone aging issue. Nature 421, 776 (2003) (link to article)
Gurdon, J. B., et al. The developmental capacity of nuclei transplanted from keratinized skin cells of adult frogs. Journal of Embryology and Experimental Morphology 34, 93–112 (1975)
Meshorer, E., et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Developmental Cell 10, 105–116 (2006)
Puri, P. L., et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Molecular Cell 1, 35–45 (1997)
Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432 (2007) (link to article)
Vucic, E. A., et al. Epigenetics of cancer progression. Pharmacogenomics 9, 215–234 (2008)
White, K. P., et al. Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science 276, 114–117 (1997) doi:10.1126/science.276.5309.114
White, K. P., et al. Microarray analysis of Drosophila development during metamorphosis. Science 286, 2179–2184 (1999) doi:10.1126/science.286.5447.2179
Wilmut, I., et al. Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813 (1997) doi:10.1038/385810a0 (link to article)



 Figure 3: Epigenetic gene regulation during mammalian development.
Figure 3: Epigenetic gene regulation during mammalian development.


























