« Prev Next »
Charles Darwin and Alfred Wallace (1858) famously proposed that positive selection could explain the many marvelous adaptations that suit organisms to their environments and lifestyles, and this simple process remains the central explanation for all evolutionary adaptation yet today. Positive selection is by no means the only component of evolution, however. In humans, at least, the great majority of mutations are thought to be selectively neutral, conferring neither benefit nor cost on their bearers (Hellmann et al., 2003). The frequency of some of these neutral genetic variants (alleles) increases simply by chance, and the resulting "genetic drift" is thought to be the most common process in human evolution (Kimura, 1968). Moreover, when selection does occur, it is most often in the form of negative, or purifying, selection, which removes new deleterious mutations as they arise, rather than promoting the spread of new traits (Kreitman, 2000).
Advantageous Alleles and Selective Sweep
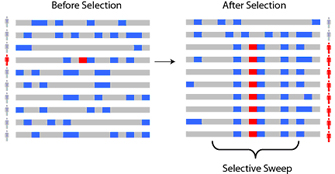
As advantageous alleles that are under positive selection increase in prevalence, these alleles leave distinctive signatures, or patterns of genetic variation, in the DNA sequence. Consider a population of individuals for which, before selection, there are hundreds of thousands of varied chromosomes in the population, all with different combinations of genetic variants. Now, say that an advantageous allele arises as a mutation on one copy of a chromosome. Through succeeding generations, the descendants of this copy, including the selected allele and nearby "hitchhiking" alleles, become more and more common through a process called a "selective sweep" (Figure 1). Note that the entire chromosome is not passed down as a unit, however; rather, because of recombination, segments of the chromosome are inherited. Thus, while the selected allele and hitchhiking alleles increase in prevalence in a selective sweep, at the same time, the segment that includes the selected allele is slowly reduced in size by recombination. Investigators are interested in the types of signals that can be detected in a selective sweep, as well as their properties and technical challenges (Nielsen, 2005; Sabeti et al., 2006).
Evidence of Positive Selection in Humans
Within the last decade, our ability to probe our own species for evidence of selection has increased dramatically due to the flood of genetic data that have been generated. Starting with the complete sequence of the human genome (Lander et al., 2001), which provides a framework and standard reference for all human genetics, key data sets include the completed or near-completed genomes of several related species (e.g., chimpanzee, macaque, gorilla, and orangutan), a public database of known genetic variants in humans, and surveys of genetic variation in hundreds of individuals in multiple populations (Chimpanzee Sequencing and Analysis Consortium, 2005; Gibbs et al., 2007; Sherry et al., 2001; International HapMap Consortium, 2007). With these new data, it is now possible to scan the entire human genome in search of signals of natural selection.
Although the study of natural selection in humans is still in an early stage, the new data, building on decades of earlier work, are beginning to reveal some of the landscape of selection in our species. In fact, researchers have identified many genetic loci at which selection has likely occurred, and some of the selective pressures involved have been elucidated. Three significant forces that have been identified thus far include changes in diet, changes in climate, and infectious disease.
Lactose Tolerance
The domestication of plants and animals roughly 10,000 years ago profoundly changed human diets, and it gave those individuals who could best digest the new foods a selective advantage. The best understood of these adaptations is lactose tolerance (Sabeti et al., 2006; Bersaglieri et al., 2004). The ability to digest lactose, a sugar found in milk, usually disappears before adulthood in mammals, and the same is true in most human populations. However, for some people, including a large fraction of individuals of European descent, the ability to break down lactose persists because of a mutation in the lactase gene (LCT). This suggests that the allele became common in Europe because of increased nutrition from cow's milk, which became available after the domestication of cattle. This hypothesis was eventually confirmed by Todd Bersaglieri and his colleagues, who demonstrated that the lactase persistence allele is common in Europeans (nearly 80% of people of European descent carry this allele), and it has evidence of a selective sweep spanning roughly 1 million base pairs (1 megabase). Indeed, lactose tolerance is one of the strongest signals of selection seen anywhere in the genome. Sarah Tishkoff and colleagues subsequently found a distinct LCT mutation also conferring lactose tolerance, in this case in African pastoralist populations, suggesting the action of convergent evolution (Tishkoff et al., 2007).
Malaria Resistance
Malaria's power to drive selection is not surprising, as it is one of the human population's oldest diseases and remains one of the greatest causes of morbidity and mortality in the world today, infecting hundreds of millions of people and killing 1 to 2 million children in Africa each year. In fact, malaria was responsible for the first case of positive selection demonstrated genetically in humans. In the 1940s and 1950s, J. B. S. Haldane and A. C. Allison demonstrated that the geographical distribution of the sickle-cell mutation (Glu6Val) in the beta hemoglobin gene (HBB) was limited to Africa and correlated with malaria endemicity, and that individuals who carry the sickle-cell trait are resistant to malaria (Allison, 1954). Since then, many more alleles for malaria resistance have shown evidence of selection, including more mutations in HBB, as well as mutations causing other red blood cell disorders (e.g., a-thalassemia, G6PD deficiency, and ovalocytosis) (Kwiatkowski, 2005).
Malaria also drove one of the most striking genetic differences between populations. This difference involves the Duffy antigen gene (FY), which encodes a membrane protein used by the Plasmodium vivax malaria parasite to enter red blood cells, a critical first step in its life cycle. A mutation in FY that disrupts the protein, thus conferring protection against P. vivax malaria, is at a frequency of 100% throughout most of sub-Saharan Africa and virtually absent elsewhere; such an extreme difference in allele frequency is very rare for humans.
Pigmentation
The role of selection in controlling human pigmentation is not a new idea; in fact, it was first advanced by William Wells in 1813, long before Darwin's formulation of natural selection (Wells, 1818). In recent years, signals of positive selection have been identified in many genes, with some signals solely in Europeans, some solely in Asians, and some shared across both continents (Lao et al., 2007; McEvoy et al., 2006; Williamson et al., 2007). Evidence for purifying selection has also been found to maintain dark skin color in Africa, where sunlight exposure is great.
A good example of selection for lighter pigmentation is the gene SLC24A5, which was one of the first to be characterized. Rebecca Lamason and her colleagues identified a mutation in the zebrafish homologue of this gene that is responsible for pigmentation phenotype. The investigators then demonstrated that a human variant in the gene explains roughly one-third of the variation in pigmentation between Europeans and West Africans, and that the European variant had likely been a target of selection (Lamason et al., 2005). In related work, Angela Hancock and her colleagues examined many genes involved in metabolism, and they showed that alleles of these genes show evidence of positive selection and correlate strongly with climate, suggesting that humans adapted to cooler climates by changing their metabolic rates (Hancock et al., 2008).
Signals for Positive Selection Mark the First Step to Understanding the Story of These Loci
While these instances of selection illustrate the power this line of research has to answer important biological and historical questions, in most cases, little or nothing of the underlying story is understood. For the great majority of selective sweeps, the pressure that drove selection, the trait selected for, and even the specific gene involved are unknown. Understanding these will require case-by-case study, identifying the possible causal mutations within each region based on strength of signal and function (e.g., mutations that alter amino acids or gene regulatory regions), and then finding the biological effects of each.
Such detailed investigations are underway, and they are intriguing. For example, a strong signal of selection in Asia localizes to amino acid substitution in the gene EDAR Sabeti et al., 2007). Mutations in EDAR cause defects in the development of hair, teeth, and exocrine glands in both mice and humans. Meanwhile, there is also evidence for selection at other genes in the same pathway in humans, as well as in stickleback fish (Colosimo et al., 2005), where the pathway regulates scale development. The phenotypic variation for this mutation is only just being elucidated, but it has already been linked to thicker head hair in Asia and has been shown to affect gene activity in the molecular pathway (Bryk et al., 2008; Fujimoto et al., 2008), although what trait was actually under selection is not yet clear. In another case, Scott Williamson and his colleagues found the strongest signal of selection in Europe and Asia at the gene DTNA, a component of the dystrophin complex (Williamson et al., 2007). While the target polymorphism and genetic variation have yet to be elucidated, the dystrophin complex is known to be important in the architecture of muscle tissue, as well as in the pathogenesis of many infectious agents, including arenaviruses and mycobacterium leprae. Another candidate gene for selection, LARGE, is also important for dystrophin function, and it has been shown to be critical for entry of various arenaviruses, including Lassa virus (Sabeti et al., 2007).
Understanding the biology behind these cases, and the many others like them, will not be easy, and it will require contributions from diverse fields, including genetics, molecular biology, developmental biology, and the study of model organisms (Figure 2). Nevertheless, the potential rewards are high. Through the study of natural selection in humans, researchers hope to learn more about how our species has changed over time, about the challenges the species has faced and how it has overcome them, and about past and present causes of disease.
References and Recommended Reading
Allison, A. C. Protection afforded by sickle-cell trait against subtertian malareal infection. British Medical Journal 4857, 290–294 (1954)
Bersaglieri, T., et al. Genetic signatures of strong recent positive selection at the lactase gene. American Journal of Human Genetics 74, 1111–1120 (2004)
Bryk, J., et al. Positive selection in East Asians for an EDAR allele that enhances NF-kappaB activation. PLoS ONE 3, e2209 (2008) (link to article)
Chimpanzee Sequencing and Analysis Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437, 69–87 (2005) doi:10.1038/nature04072 (link to article)
Colosimo, P. F., et al. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307, 1928–1933 (2005)
Darwin, C. & Wallace, A. R. On the tendency of species for form varieties; and on the perpetuation of varieties and species by natural means of selection. Proceedings of the Linnean Society of London 3, 45–62 (1858)
Fujimoto, A., et al. A scan for genetic determinants of human hair morphology: EDAR is associated with Asian hair thickness. Human Molecular Genetics 17, 835–843 (2008)
Gibbs, R. A,. et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316, 222–234 (2007)
Hancock, A. M., et al. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genetics 4, e32 (2008) (link to article)
Hellmann, I., et al. A neutral explanation for the correlation of diversity with recombination rates in humans. American Journal of Human Genetics 72, 1527–1535 (2003)
International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature 449, 851–861 (2007) doi:10.1038/nature06258 (link to article)
Kimura, M. Evolutionary rate at the molecular level. Nature 217, 624–626 (1968) doi:10.1038/217624a0 (link to article)
Kreitman, M. Methods to detect selection in populations with applications to the human. Annual Review of Genomics and Human Genetics 1, 539–559 (2000)
Kwiatkowski, D. P. How malaria has affected the human genome and what human genetics can teach us about malaria. American Journal of Human Genetics 77, 171–192 (2005)
Lamason, R. L., et al. SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310, 1782–1786 (2005)
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001) doi:10.1038/ 409860a0 (link to article)
Lao, O., et al. Signatures of positive selection in genes associated with human skin pigmentation as revealed from analyses of single nucleotide polymorphisms. Annals of Human Genetics 71, 354–369 (2007)
McEvoy, B., et al. The genetic architecture of normal variation in human pigmentation: An evolutionary perspective and model. Human Molecular Genetics 15, R176–181 (2006)
Nielsen, R. Molecular signatures of natural selection. Annual Review of Genetics 39, 197–218 (2005)
Sabeti, P. C., et al. Positive natural selection in the human lineage. Science 312, 1614–1620 (2006)
Sabeti, P. C., et al. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918 (2007) doi:10.1038/nature06250 (link to article)
Sherry, S. T., et al. dbSNP: The NCBI database of genetic variation. Nucleic Acids Research 29, 308–311 (2001)
Tishkoff, S. A., et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nature Genetics 39, 31–40 (2007) doi:10.1038/ng1946 (link to article)
Wells, W. C. Two Essays: One Upon Single Vision with Two Eyes; The Other on Dew (Edinburgh, Constable, 1818)
Williamson, S. H., et al. Localizing recent adaptive evolution in the human genome. PLoS Genetics 3, e90 (2007) (link to article)
World Health Organization. WHO expert committee on malaria. World Health Organization Technical Report Series 892, 1–74 (2000)




 Figure 2
Figure 2


























