« Prev Next »
The external environment's effects upon genes can influence disease, and some of these effects can be inherited in humans. Studies investigating how environmental factors impact the genetics of an individual's offspring are difficult to design. However, in certain parts of the world in which social systems are highly centralized, environmental information that might have influenced families can be obtained. For example, Swedish scientists recently conducted investigations examining whether nutrition affected the death rate associated with cardiovascular disease and diabetes and whether these effects were passed from parents to their children and grandchildren (Kaati et al., 2002). These researchers estimated how much access individuals had to food by examining records of annual harvests and food prices in Sweden across three generations of families, starting as far back as the 1890s. These researchers found that if a father did not have enough food available to him during a critical period in his development just before puberty, his sons were less likely to die from cardiovascular disease. Remarkably, death related to diabetes increased for children if food was plentiful during this critical period for the paternal grandfather, but it decreased when excess food was available to the father. These findings suggest that diet can cause changes to genes that are passed down though generations by the males in a family, and that these alterations can affect susceptibility to certain diseases. But what are these changes, and how are they remembered? The answers to questions such as these lie in the concept of epigenetics.
What Is Epigenetics? How Do Epigenetic Changes Affect Genes?
Epigenetics involves genetic control by factors other than an individual's DNA sequence. Epigenetic changes can switch genes on or off and determine which proteins are transcribed.
Epigenetics is involved in many normal cellular processes. Consider the fact that our cells all have the same DNA, but our bodies contain many different types of cells: neurons, liver cells, pancreatic cells, inflammatory cells, and others. How can this be? In short, cells, tissues, and organs differ because they have certain sets of genes that are "turned on" or expressed, as well as other sets that are "turned off" or inhibited. Epigenetic silencing is one way to turn genes off, and it can contribute to differential expression. Silencing might also explain, in part, why genetic twins are not phenotypically identical. In addition, epigenetics is important for X-chromosome inactivation in female mammals, which is necessary so that females do not have twice the number of X-chromosome gene products as males (Egger et al., 2004). Thus, the significance of turning genes off via epigenetic changes is readily apparent.
Within cells, there are three systems that can interact with each other to silence genes: DNA methylation, histone modifications, and RNA-associated silencing (Figure 1; Egger et al., 2004).
DNA Methylation
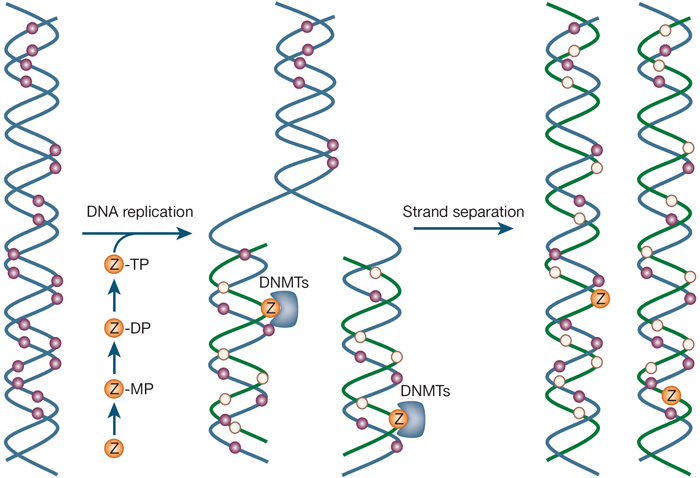
DNA methylation is a chemical process that adds a methyl group to DNA. It is highly specific and always happens in a region in which a cytosine nucleotide is located next to a guanine nucleotide that is linked by a phosphate; this is called a CpG site (Egger et al., 2004; Jones & Baylin, 2002; Robertson, 2002). CpG sites are methylated by one of three enzymes called DNA methyltransferases (DNMTs) (Egger et al., 2004; Robertson, 2002). Inserting methyl groups changes the appearance and structure of DNA, modifying a gene's interactions with the machinery within a cell's nucleus that is needed for transcription. DNA methylation is used in some genes to differentiate which gene copy is inherited from the father and which gene copy is inherited from the mother, a phenomenon known as imprinting.
Histone Modifications
Histones are proteins that are the primary components of chromatin, which is the complex of DNA and proteins that makes up chromosomes. Histones act as a spool around which DNA can wind. When histones are modified after they are translated into protein (i.e., post-translation modification), they can influence how chromatin is arranged, which, in turn, can determine whether the associated chromosomal DNA will be transcribed. If chromatin is not in a compact form, it is active, and the associated DNA can be transcribed. Conversely, if chromatin is condensed (creating a complex called heterochromatin), then it is inactive, and DNA transcription does not occur.
There are two main ways histones can be modified: acetylation and methylation. These are chemical processes that add either an acetyl or methyl group, respectively, to the amino acid lysine that is located in the histone. Acetylation is usually associated with active chromatin, while deacetylation is generally associated with heterochromatin. On the other hand, histone methylation can be a marker for both active and inactive regions of chromatin. For example, methylation of a particular lysine (K9) on a specific histone (H3) that marks silent DNA is widely distributed throughout heterochromatin. This is the type of epigenetic change that is responsible for the inactivated X chromosome of females. In contrast, methylation of a different lysine (K4) on the same histone (H3) is a marker for active genes (Egger et al., 2004).
RNA-Associated Silencing
Genes can also be turned off by RNA when it is in the form of antisense transcripts, noncoding RNAs, or RNA interference. RNA might affect gene expression by causing heterochromatin to form, or by triggering histone modifications and DNA methylation (Egger et al., 2004).
Epigenetics and Disease: Some Examples
Epigenetics and Cancer
The first human disease to be linked to epigenetics was cancer, in 1983. Researchers found that diseased tissue from patients with colorectal cancer had less DNA methylation than normal tissue from the same patients (Feinberg & Vogelstein, 1983). Because methylated genes are typically turned off, loss of DNA methylation can cause abnormally high gene activation by altering the arrangement of chromatin. On the other hand, too much methylation can undo the work of protective tumor suppressor genes.
As previously mentioned, DNA methylation occurs at CpG sites, and a majority of CpG cytosines are methylated in mammals. However, there are stretches of DNA near promoter regions that have higher concentrations of CpG sites (known as CpG islands) that are free of methylation in normal cells. These CpG islands become excessively methylated in cancer cells, thereby causing genes that should not be silenced to turn off. This abnormality is the trademark epigenetic change that occurs in tumors and happens early in the development of cancer (Egger et al., 2004; Robertson, 2002; Jones & Baylin, 2002). Hypermethylation of CpG islands can cause tumors by shutting off tumor-suppressor genes. In fact, these types of changes may be more common in human cancer than DNA sequence mutations (Figure 2).
Furthermore, although epigenetic changes do not alter the sequence of DNA, they can cause mutations. About half of the genes that cause familial or inherited forms of cancer are turned off by methylation. Most of these genes normally suppress tumor formation and help repair DNA, including O6-methylguanine-DNA methyltransferase (MGMT), MLH1 cyclin-dependent kinase inhibitor 2B (CDKN2B), and RASSF1A. For example, hypermethylation of the promoter of MGMT causes the number of G-to-A mutations to increase (Figure 2).
Hypermethylation can also lead to instability of microsatellites, which are repeated sequences of DNA. Microsatellites are common in normal individuals, and they usually consist of repeats of the dinucleotide CA. Too much methylation of the promoter of the DNA repair gene MLH1 can make a microsatellite unstable and lengthen or shorten it (Figure 2). Microsatellite instability has been linked to many cancers, including colorectal, endometrial, ovarian, and gastric cancers (Jones & Baylin, 2002).
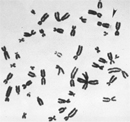
Epigenetics and Mental Retardation
Fragile X syndrome is the most frequently inherited mental disability, particularly in males. Both sexes can be affected by this condition, but because males only have one X chromosome, one fragile X will impact them more severely. Indeed, fragile X syndrome occurs in approximately 1 in 4,000 males and 1 in 8,000 females. People with this syndrome have severe intellectual disabilities, delayed verbal development, and "autistic-like" behavior (Penagarikano et al., 2007).
Fragile X syndrome gets its name from the way the part of the X chromosome that contains the gene abnormality looks under a microscope; it usually appears as if it is hanging by a thread and easily breakable (Figure 3). The syndrome is caused by an abnormality in the FMR1 (fragile X mental retardation 1) gene. People who do not have fragile X syndrome have 6 to 50 repeats of the trinucleotide CGG in their FMR1 gene. However, individuals with over 200 repeats have a full mutation, and they usually show symptoms of the syndrome. Too many CGGs cause the CpG islands at the promoter region of the FMR1 gene to become methylated; normally, they are not. This methylation turns the gene off, stopping the FMR1 gene from producing an important protein called fragile X mental retardation protein. Loss of this specific protein causes fragile X syndrome. Although a lot of attention has been given to the CGG expansion mutation as the cause of fragile X, the epigenetic change associated with FMR1 methylation is the real syndrome culprit.
Fragile X syndrome is not the only disorder associated with mental retardation that involves epigenetic changes. Other such conditions include Rubenstein-Taybi, Coffin-Lowry, Prader-Willi, Angelman, Beckwith-Wiedemann, ATR-X, and Rett syndromes (Table 1).
Combating Diseases with Epigenetic Therapy
Because so many diseases, such as cancer, involve epigenetic changes, it seems reasonable to try to counteract these modifications with epigenetic treatments. These changes seem an ideal target because they are by nature reversible, unlike DNA sequence mutations. The most popular of these treatments aim to alter either DNA methylation or histone acetylation.
Inhibitors of DNA methylation can reactivate genes that have been silenced. Two examples of these types of drugs are 5-azacytidine and 5-aza-2′-deoxycytidine (Egger et al., 2004). These medications work by acting like the nucleotide cytosine and incorporating themselves into DNA while it is replicating. After they are incorporated into DNA, the drugs block DNMT enzymes from acting, which inhibits DNA methylation.
Drugs aimed at histone modifications are called histone deacetylase (HDAC) inhibitors. HDACs are enzymes that remove the acetyl groups from DNA, which condenses chromatin and stops transcription. Blocking this process with HDAC inhibitors turns on gene expression. The most common HDAC inhibitors include phenylbutyric acid, SAHA, depsipeptide, and valproic acid (Egger et al., 2004).
Caution in using epigenetic therapy is necessary because epigenetic processes and changes are so widespread. To be successful, epigenetic treatments must be selective to irregular cells; otherwise, activating gene transcription in normal cells could make them cancerous, so the treatments could cause the very disorders they are trying to counteract. Despite this possible drawback, researchers are finding ways to specifically target abnormal cells with minimal damage to normal cells, and epigenetic therapy is beginning to look increasingly promising.
References and Recommended Reading
Egger, G., et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature 429, 457–463 (2004) doi:10.1038/nature02625 (link to article)
Feinberg, A.P., & Vogelstein, B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 301, 89–92 (1983) doi:10.1038/301089a0 (link to article)
Jones, P. A., & Baylin, S. B. The fundamental role of epigenetic events in cancer. Nature Reviews Genetics 3, 415–428 (2002) doi:10.1038/nrg816 (link to article)
Kaati, G., et al. Cardiovascular and diabetes mortality determined by nutrition during parents' and grandparents' slow growth period. European Journal of Human Genetics 10, 682–688 (2002)
Penagarikano, O., et al. The pathophysiology of fragile X syndrome. Annual Review of Genomics and Human Genetics 8, 109–129 (2007) doi:10.1146/annurev.genom.8.080706.092249
Robertson, K. D. DNA methylation and chromatin: Unraveling the tangled web. Oncogene 21, 5361–5379 (2002) doi:10.1038/sj.onc.1205609



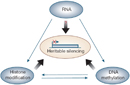
 Figure 1
Figure 1