« Prev Next »
A pile of sugar or a bottle of olive oil can sit unchanged for years at room temperature. Yet at similar temperatures, cells can convert the molecules that compose sugar and oil into a vast array of other molecules, and release energy at the same time. Typically, chemical reactions require high temperatures, strong acids or bases, or unusual solvents. How do enzymes manage to perform conversion reactions without these chemical tools? The key components in these cellular reactions are enzymes — protein catalysts that speed up reactions, specify the products that are made, and allow for careful control of the reaction rates. How did scientists figure out these essential reaction-facilitating features of enzymes? The story of research on serine proteases provides an example.
Enzymes Are Protein Catalysts
A catalyst is a molecule that increases the rate of a chemical reaction without being consumed by the reaction. When the reaction is finished, the amount of catalyst has not changed. However, a chemical reaction accelerated by a catalyst must still be energetically downhill, if not it would not occur. The reactions that convert sucrose or olive oil, in the presence of oxygen, into carbon dioxide and water, and release large amounts of free energy. However, the rates at which these energetically downhill conversions take place at room temperature are essentially zero, as the molecules of sugar and olive oil are kinetically stable. The key reactions in cell metabolism are catalyzed by enzymes, which allow some desired reactions to happen quickly while not changing the rates of other reactions. How do they do this?
Proteases Are Common and Widespread Enzymes
Proteases are enzymes that break the peptide bond that joins amino acids together in proteins. They are examples of hydrolases, enzymes that break a chemical bond by the addition of a water molecule. Although the hydrolysis of a peptide bond is energetically downhill, this process is very slow at normal temperature and pH. A group of chemists measured this rate, and found that a simple peptide bond between two amino acids in water and room temperature has a half-life of several years. (Kahne & Still 1988) For example, hair and wool, which are composed almost entirely of protein, can remain in water for weeks and not break down. Similarly, protein-rich foods can be boiled for hours and not break down into amino acids. Yet these same foods, in our digestive systems, can be completely separated into amino acids within hours. How do enzymes do this?
Serine Proteases
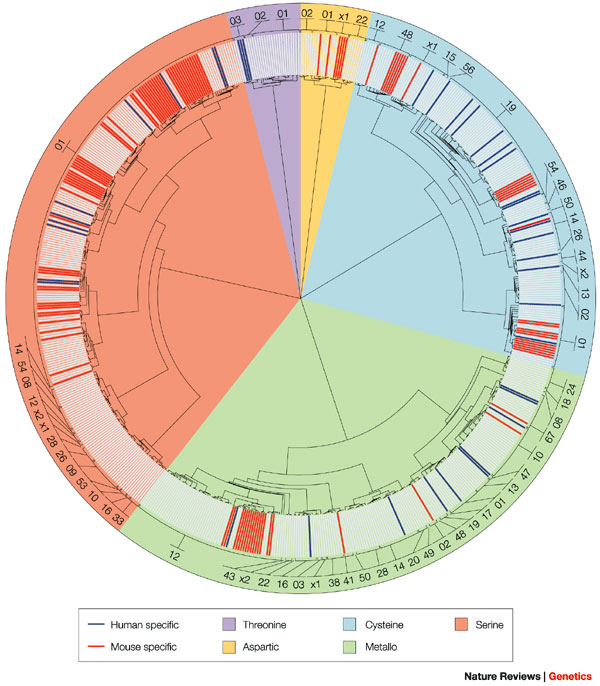
Serine proteases are involved in an enormous number of biological processes (Stroud 1974). In every type of organism that scientists examine, they continue to discover new enzymes in this group. These enzymes are powerful. They digest our food, help our blood clot, fight infections, and help sperm enter eggs. They help bacteria digest material (e.g., "flesh eating" Staph) and help viruses infect cells (hepatitis C). They also regulate the development of organisms and degrade flesh after a snake bite. (Rose & Di Cera 2000). In humans, they are prevalent in many physiological functions, both normal and disease-related (Figure 1) (Puente et al. 2003).
Chymotrypsin as a Model Enzyme
The first serine protease scientists explored in detail was chymotrypsin, a digestive enzyme synthesized in our pancreas, along with the related enzymes trypsin and elastase. It is transported to our small intestine in an inactive form. Once the enzyme enters our small intestine, it becomes activated. Purifying it was not difficult for scientists, and its structure is quite simple, with only a single subunit and a single active site per molecule. For those reasons, it became an easy resource for studying how enzymes in general work. Once researchers were able to isolate high quality chymotrypsin crystals, they were then able to precisely locate very important amino acids in its three dimensional structure (more about this below). So far, researchers have identified at least 300 enzymes that use the same mechanism as chymotrypsin (Rawlings et al. 2008). As you might expect from their name, the mechanism has something to do with the amino acid serine.
How Do We Know Serine Is Involved?
A powerful tool scientists can use to figure out what an enzyme does is an inhibitor. One group of inhibitors, affinity labeling reagents, resemble normal substrates but have the useful property of forming stable covalent bonds with the enzymes they ‘poison'. Since these molecules block the enzyme from working, scientists can use these molecules to identify specific amino acids that are important for the enzyme's function. The chemical diisopropyl fluorophosphate, (DFP), a poison known as a nerve gas, forms a stable bond with the side chain oxygen of specific serines in enzymes. When scientists treated chymotrypsin with DFP, and identified the amino acids around the modified serine, they found that only a single serine, serine 195, reacted (Dixon & Neurath 1956). (The numbering convention for amino acids in proteins is that the amino-terminal amino acid is numbered 1, and they are then sequentially numbered towards the other end, the carboxyl terminus). When this experiment was repeated with similar enzymes such as trypsin and elastase, they again found that a single serine reacted and that the amino acids surrounding this serine were almost identical in all of the enzymes they tested. When they put this evidence together, they found that this single serine was much more reactive than any of the other 26 amino acids present in the protein. Blocking this serine permanently destroyed the function of the enzyme. Moreover, the sequences around this amino acid were constant in several different enzymes. Putting these pieces together, Dixon & Neurath concluded that the side chain of this serine must be an important component of the enzymatic action. In similar work, researchers found that another inhibitor, TPCK (N-tosyl l-phenylalanyl chloromethyl ketone), specifically reacted with one histidine, histidine 57, in chymotrypsin (Schoellmann & Shaw 1963).
A Covalent Intermediate Is Formed in the Reaction of Chymotrypsin
Hartley & Kilby (1954) studied the reaction of chymotrypsin with the substrate p-nitrophenyl acetate. This molecule resembles the normal protein substrates of chymotrypsin, but when the enzyme reacts it produces a yellow color easy to follow in the laboratory. They found that the reaction of this substrate occurred in two steps. In the first step, the p-nitrophenyl group is released quickly but the acetate group remains covalently bound to the enzyme. In a second, slower step, the enzyme reacts with water and the acetate group is broken away from the enzyme. This second step was necessary before the enzyme could repeat a reaction cycle.
Crystals Clarify the Picture and Suggest a Mechanism
X-ray crystallography is an important tool for studying the 3-dimensional structure of proteins. A research group in England used this method to work out the structure of chymotrypsin. When they carefully examined the structure of chymotrypsin, they found the histidine and serine amino acids described above were close to each other in the folded protein structure, even though they were not close together in the primary sequence of the protein. In particular, the imidazole side chain of histidine 57 was very close to the hydroxyl group of serine 195, close enough to form a hydrogen bond. Aspartic acid 102 was also close to the side chain of histidine, but on the opposite side from the serine (Figure 2) (Blow et al. 1969).
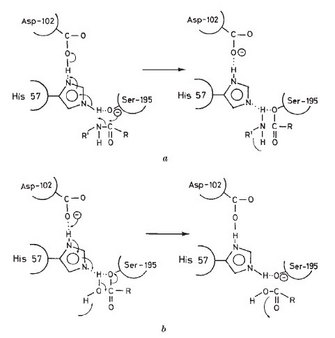
These scientists suggested that this arrangement would allow the movement of protons and charge in the active site of the enzyme. The histidine was in position to act as a base, a proton acceptor, and remove the proton from the OH group of serine. With this change, the serine is much more reactive, and can easily form a new bond with the carbon atom in the peptide bond of the substrate. The negative charge of the carboxylic acid group of the aspartic acid would then be in the correct position to stabilize the positively charged side chain of the histidine. This specific spatial arrangement of aspartic acid, histidine, and serine has been found in hundreds of enzymes and is often referred to as "the catalytic triad" (Blow et al. 1969.) An excellent review of the roles of acids and bases in biology can be found in Allen (2010), and the overall sequence of the reactions is shown in Stroud (1974).
What Common Structural Features Are Found in These Enzymes?
As scientists studied the structures of large numbers of serine proteases they discovered several common structural features. They found specific aspartic acid, histidine, and serine amino acids in a special alignment that allowed the easy transfer of protons into and out of the active site. They found a pocket lined with charged groups that would stabilize the intermediate steps of making a breaking peptide and ester bond (the "oxyanion hole"). They also found a much more variable region which was the basis for the specificity of the enzyme.
This 3-D arrangement of amino acids optimized to catalyze peptide bond hydrolysis has clearly developed at least twice in evolution, an example of convergent evolution at the molecular level. (Brenner 1988) The bacterial protease subtilisin uses all of the catalytic mechanisms seen for chymotrypsin, yet its amino acid sequence is very different. Although the folded tertiary structures of the bacterial and mammalian enzymes have some differences, they do share the same arrangement of the catalytic triad amino acids. We now understand the mechanism and amino acid sequences shared by serine proteases well enough that we can identify new serine proteases by examining their amino acid sequence.
What Features Allow Different Serine Proteases to Attack Different Substrates?
What is different in each of the serine proteases examined is the specificity pocket. In chymotrypsin, this pocket is lined with hydrophobic amino acids, so substrate proteins with hydrophobic amino acids, such as leucine or isoleucine, bind tightly and in the correct orientation for the catalytic triad to function. On the other hand, the pocket for trypsin has a negatively charged aspartic acid in the pocket, so the substrates that trypsin breaks apart must have a positively charged amino acid, such as lysine or arginine, in the proper position. As another example, in the enzyme elastase, this pocket is very small, so only proteins with relatively small amino acid side chains, such as glycine or alanine, can be cut by this enzyme. These are digestive enzymes capable of cutting peptide bonds in a wide range of proteins. In some pathways, such as blood clotting or the immune system, a serine protease may be so specific that it only can cut a single peptide bond in a single unique protein substrate.
Planned Mutations with Surprising Results
The ability to manipulate the DNA sequences of genes has given scientists another tool to figure out the roles of specific amino acids in enzymes. By site-directed mutagenesis, a desired amino acid change can be made in the gene for the protein being studied. When this mutated gene is used to make a new protein, the result of changing the amino acid can be examined. Scientists have done this with serine proteases, and some have used this technique to make targeted changes in the amino acids lining the specificity pocket of trypsin (Craik et al. 1985). They were surprised to find that altering single amino acids in the specificity pocket changed the specificity, as they expected, but at the same time altered the overall catalytic properties of the enzyme. They found the maximum catalytic rates of the altered enzymes were decreased by factors of 10 to a 1000 compared to the original enzyme. Scientists think that this is a general property of many enzymes. Although we can identify specific amino acids as being involved in the specific chemical reactions of catalysis, the ability of the protein to work as an effective catalyst may also rely on amino acid interactions far away from the active site.
How Might an Enzyme Mechanism Like This Arise?
As a protein like a serine protease evolves, how might the earliest versions have worked, before the current elegant enzyme structures had evolved? Scientists used targeted mutations in the protease subtilisin to explore some possibilities (Carter & Wells 1988). They changed the reactive serine (serine 221) in subtilisin to alanine. This mutated enzyme still bound substrates, but they found its ability to act as a catalyst was reduced by a factor of a million. However, they did find that this mutated version of subtilisin still acted as a catalyst, and would break peptide bonds 1000 times faster than would occur with no catalyst. Thus even earlier forms of these enzymes, those that existed before the catalytic triad mechanism had evolved into their structure, would still be valuable to the organisms in which they had originally evolved.
The result of these mutation experiments also changed scientists' view of what features were important in this group of enzymes. Clearly the catalytic triad of aspartic acid, serine, and histidine is important, but other portions of the enzyme are also important in the catalytic process. Also, our ability to fully predict how the three dimensional structure of a protein will change after a single amino acid substitution takes place is clearly far from perfect. The ability to predict how specific amino acid changes influence larger scale protein structure is one of the most difficult challenges in biochemistry and cell biology
Not All Proteases Are Serine Proteases
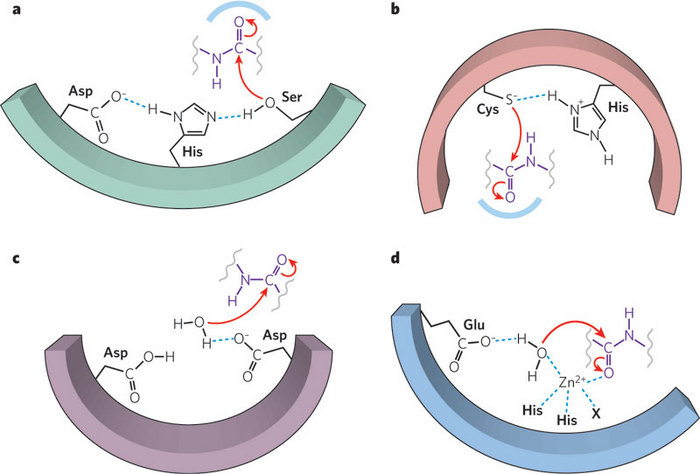
There are other large groups of proteases that share common mechanisms, but do not use an active serine. For example, there are cysteine peptidases, such as papain from papaya, in which the sulfur in cysteine acts much like the oxygen on serine in serine proteases. There are also aspartic acid peptidases, which include HIV protease, an important target for drug development. There are a wide range of metalloproteases that use the interaction of a specific metal ion to help the reaction (Figure 3). And there are proteases for which scientists have not yet discovered the mechanism of action.
Do Enzyme Mechanisms Have Any Features in Common?
No single mechanism can explain the wealth of chemical reactions carried out by biological enzymes. They can transfer electrons and protons, assemble small molecules into large molecules and break them back into smaller pieces, accurately copy DNA, and repair DNA when it is damaged. Above all these specific functions, there is indeed one commonality among all enzymes; they carry out reactions identical to the types of reactions carried out by organic chemists. Over the course of evolution, enzymes have become astounding catalysts. In the words of one prominent scientist, enzymes are not different, they are just better (Knowles 1991).
Summary
Scientists have combined data from chemical labeling studies, X-ray crystallography, and planned mutations to gain an understanding of how serine proteases can accelerate the rate of peptide bond hydrolysis. They have been able to identify specific amino acids involved in the catalytic steps used by these enzymes. They have also been able to recreate enzymes that may have been intermediate versions of these enzymes during evolution. However, we are still unable to fully predict how small amino acid changes can alter the structure and function of proteins. Scientists are continuing to discover new members of this diverse protein family, which is vital to the life of all organisms.
References and Recommended Reading
Allen, K. 2010 Acids and Bases. Nature Chemical Biology 6, June 2010 doi: 10.1038/nchembio.382.
Blow, D. M., Birkthoft, J. J. et al. Role of a buried acid group in the mechanism of action of chymotrypsin. Nature 221, 337–340 (1969). doi:10.1038/221337a0
Brenner, S. The molecular evolution of genes and proteins: a tale of two serines. Nature 334, 528–530 (1988). doi:10.1038/334528a0
Carter, P. & Wells, J. A. Dissecting the catalytic triad of a serine protease. Nature 332, 564–568 (1988). doi:10.1038/332564a0
Craik, C. S., Largman, C. et al. Redesigning trypsin: alteration of substrate specificity. Science 237, 291–297 (1985). doi: 10.1126/science.3838593
Delbaere, L. T. J., Hutcheon, W. L. B. et al. Tertiary structural differences between microbial serine proteases and pancreatic serine enzymes. Nature 257, 758–763 (1975). doi:10.1038/257758a0
Dixon, G. H. & Neurath, H. Biochimica et Biophysica Acta 20, 572–274 (1956).
Erez, E., Fass, D. et al. How intramembrane proteases bury hydrolytic reactions in the membrane. Nature 459, 371–378 (2009). doi:10.1038/nature08146
Hartly, B. S. & Kilby, B. A. The reaction of p-nitrophenyl esters with chymotrypsin and insulin. Biochemical Journal 56, 288–297 (1954).
Kahne, D. & Still, W. C. Hydrolysis of a peptide bond in neutral water. Journal of the American Chemical Association 110, 7529–7534 (1988).
Knowles, J. Enzyme catalysis: not different, just better. Nature 350, 121–124 (1991). doi:10.1038/350121a0
Puente, X. S., Sánchez, L. M. et al. Human and mouse proteases: a comparative genomic approach. Nature Reviews Genetics 4, 544–558 (2003). doi:10.1038/nrg1111
Rawlings, N. D., Morton, F. R. et al. MEROPS: The Protease Database. Nucleic Acids Research 36 Database issue, D320–325 (2008) (accessed August 24, 2010).
Rose, T. & Di Cera, E. Serine Protease Home Page. (2000) (accessed August 24, 2010).
Schoellmann, G. & Shaw E. Direct evidence for the presence of histidine in the active center of chymotrypsin. Biochemistry 2, 252–255 (1963). doi: 10.1021/bi00902a008
Stroud, R. M. A family of protein cutting enzymes. Scientific American 231, 74–88 (1974).






























