« Prev Next »
Cytogenetic approaches to studying chromosomes and their relationship to human disease have improved greatly over the past several decades. Modern cytogenetic approaches enable researchers to do the following, among other things:
- Precisely label the chromosomal location of any gene using different colored dots
- Examine cells from any type of tissue, even tumor cells
- Identify cells that have lost or gained a specific chromosome, undergone a translocation event involving a specific set of chromosomes, or lost or gained a copy of a given gene or genes
- Determine whether specific regions of chromosomes have been lost or gained without ever looking at the chromosomes under a microscope
Clearly, the field of cytogenetics has developed into a vital tool for studying and diagnosing human disease. But how did this field first emerge, and how did researchers develop the many different cytogenetic techniques that currently exist?
The Emergence of a New Field
The field of human cytogenetics was initiated in 1956, when the number of chromosomes in a diploid human cell was accurately determined to be 46 (Tjio & Levan, 1956). Since then, our knowledge of human cytogenetics and our ability to utilize cytogenetic data to understand and diagnose human disease has increased by leaps and bounds (Speicher & Carter, 2005; Trask, 2002).
As the field of human cytogenetics emerged, researchers began to develop methods to visualize chromosome structure and organization. Scientists quickly realized that not all chromosomes are created equal--specifically, they differ in their length and in the position of their centromere. Researchers also embarked on numerous studies to determine the relationship between human disease and chromosomes.
Early cytogenetic studies showed that an extra or missing copy of certain human chromosomes could lead to disease. For example, in 1959, an extra copy of chromosome 21 was shown to be associated with Down syndrome (also called trisomy 21) (Lejeune et al., 1959). In the same year, several abnormalities in sex chromosome number were linked to disease. In particular, Turner's syndrome was shown to be associated with the presence a single X chromosome and no Y chromosome (45,X) (Ford et al., 1959), whereas Klinefelter's syndrome was determined to be associated with the presence of two copies of the X chromosome and one copy of the Y chromosome (47,XXY) (Jacobs & Strong, 1959). Both Turner's syndrome and Klinefelter's syndrome affect sexual differentiation in affected invidivuals.
The Philadelphia Chromosome
Soon after discovering the link between chromosome number and disease, researchers turned their attention to the role of chromosome structure. Thus, in 1960, a collaborative study between Peter Nowell, a new faculty member at the University of Pennsylvania, and David Hungerford, a graduate student at the Institute for Cancer Research in Philadelphia, uncovered a link between chronic myelogenous leukemia (CML) and abnormal chromosome structure. Specifically, the researchers discovered the presence of a small chromosome, which they named the "Philadelphia chromosome," that was unique to CML cells (Nowell & Hungerford, 1960). Later, in 1968, Janet Rowley used new chromosome staining techniques to show that the Philadelphia chromosome arose as a result of a translocation event involving chromosomes 9 and 22 (Rowley, 1973). Then, in 1985, the breakpoint of the translocation was mapped and shown to result in the fusion of parts of the BCR gene from chromosome 22 and the ABL1 gene from chromosome 9, resulting in a gene fusion product called BCR-ABL (Heisterkamp et al., 1985). The ABL1 half of the encoded fusion protein exhibits high tyrosine kinase activity, which is largely responsible for CML-associated phenotypes. Using this information, scientists developed a drug called imatinib mesylate (also called STI571 and Gleevec) to inhibit ABL1 kinase activity (Druker, 2002). Imatinib treatment of CML has been heralded as one of the biggest success stories in cancer treatment over the past decade.
Cri du Chat Syndrome and Retinoblastoma
During the period in which Nowell and Hungerford discovered the Philadelphia chromosome, other scientists were examining the link between chromosomal deletions and disease. An important breakthrough occurred in 1963, when cri du chat syndrome, a congenital disorder that is associated with severe mental retardation and a cat-like cry in affected infants, was found to result from a loss of part of the short arm of chromosome 5 (Lejeune et al., 1963). That same year, early studies of cells from patients with retinoblastoma, a childhood form of retinal cancer, also showed deletion of a specific chromosomal region (Lele et al., 1963). This region was later determined to harbor the RB1 gene, the first official tumor suppressor gene ever identified. Studies of RB1-associated retinoblastoma led to the establishment of the two-hit hypothesis, which is a cornerstone of cancer biology (Knudson, 1971).
Using Cytogenetic Approaches to Map Genes
In addition to providing associations between chromosomal abnormalities and disease, cytogenetic approaches have also allowed researchers to map genes to particular chromosomes. For example, in 1968, Roger Donahue used new methods to study metaphase chromosomes in his own blood cells, and he noted that one of his copies of chromosome 1 had a region near the centromere that was loosely structured and uncoiled. Using his extended family pedigree and conducting biochemical tests to determine blood group markers, Donahue employed cytogenetic techniques to map the Duffy blood group locus to chromosome 1 (Donahue et al., 1968).
Shortly after the Duffy blood group locus was mapped, Maximo Drets and Margery Shaw established methods to stain metaphase chromosomes using a dye called Giemsa, which produces a signature banding pattern, called G-bands, for each of the 24 different human chromosomes (Drets & Shaw, 1971). G-banding patterns can be used to detect chromosomal translocations, deletions, and insertions, and they have made key advances in gene discovery possible. For instance, as previously mentioned, Rowley used G-banding patterns to determine that a translocation event involving chromosomes 9 and 22 was responsible for CML (Rowley, 1973). G-banding methods continue to be widely used today, though such approaches have certain drawbacks. For instance, G-banding requires metaphase chromosomes, which are easily obtained from blood samples but are more difficult to retrieve from solid tissue samples. Furthermore, metaphase chromosomes are highly condensed, which can lead to lower resolution in mapping.
Human-Mouse Somatic Cell Hybrids
Although cytogenetic approaches evolved over time such that chromosomes could be easily distinguished from each other, researchers also needed ways to study individual chromosomes in more detail. In an effort to meet this need, researchers used the Sendai virus to induce fusion between a human cell and a mouse cell, resulting in a human-mouse somatic cell hybrid that contained the complete mouse genome, as well as sparse numbers of human chromosomes (Ephrussi & Weiss, 1965; Harris & Watkins, 1965) . An extensive series of human-mouse hybrid cell lines that carried known combinations of human chromosomes was thus developed, and this series greatly facilitated the mapping of human genes to specific chromosomes prior to the advent of the Human Genome Project.
Using Flow Cytometry to Sort Chromosomes
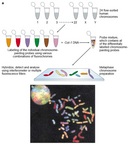
Although flow cytometry and FACS were initially used to isolate populations of intact cells, researchers eventually adapted these techniques to isolate individual human chromosomes, as shown in Figure 1. Such techniques involve using mitotic cell suspensions and disrupting the cell membranes to release the condensed chromosomes that are labeled using two different types of fluorescent dyes (Carrano et al., 1979). The first dye, called Hoechst 33269, binds to A-T base pairs, and the second dye, called chromomycin A, binds to G-C base pairs.
FACS is used to isolate individual fluorescently labeled chromosomes in solution, and with the exception of four chromosomes (9, 10, 11, and 12), all of the human chromosomes can be resolved based on their laser light-scattering properties. In addition, this approach can also be used to determine changes in chromosome size and number. Larger chromosomes contain a higher number of A-T or G-C base pairs, leading to higher levels of Hoechst and chromomycin staining, respectively. As illustrated in Figure 1, chromosome 21, which is the smallest human chromosome, shows the lowest Hoechst and chromomycin staining intensity. Chromosomes 1 and 2, which are the largest human chromosomes, show the highest Hoechst and chromomycin staining. In general, the A-T and G-C contents of a given chromosome are quite similar, and that is why a diagonal line of chromosomes, each of increasing size, connects the smallest (21) to the largest (1 and 2) in Figure 1.
Fluorescence In Situ Hybridization (FISH)
In further chromosomal studies, researchers used restriction enzymes to cut pooled chromosome populations into smaller DNA fragments. The resulting DNA fragments were ligated into DNA plasmid vectors, which allowed them to be propagated in bacteria or yeast host cells. This led to the generation of chromosome-specific collections of DNA fragments, called libraries, which served as a platform for the Human Genome Project. Furthermore, the ability to isolate collections of DNA fragments that span individual chromosomes led to the development of chromosome-specific staining methods.
Researchers also wanted to study regions of individual chromosomes within the nucleus of intact cells. Thus, they used a cytogenetic method called fluorescence in situ hybridization (FISH) to map DNA sequences to specific regions of human chromosomes. FISH involves the use of fluorescently labeled DNA probes that are capable of hybridizing to complementary chromosomal regions. This technique allows researchers to view the chromosomal location of a particular gene or DNA sequence through a microscope; the net result is a fluorescent dot at the chromosomal location where the labeled probe binds. The first single-copy human gene to be mapped using FISH was thyroglobulin in 1985 (Landegent et al., 1985). FISH allows a higher level of resolution than standard G-banding approaches.
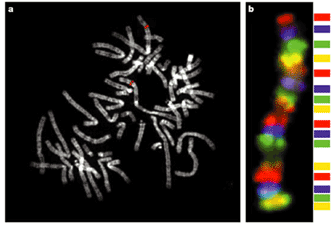
Figure 2a demonstrates an example of a standard FISH experiment, in which the red fluorescent DNA probe, corresponding to a 150-kilobase-pair region of chromosome 1, is used to label a metaphase chromosome spread. In this case, two red fluorescent dots can be observed, corresponding to the maternal and paternal copies of chromosome 1. Probes for different genes or DNA sequences can even be used simultaneously, as long as each has a different color associated with it. Figure 2b shows a FISH experiment in which a set of DNA probes that bind along the length of a human chromosome were each labeled with a different color; the net result is a rainbow-colored chromosome.
FISH has been very useful in the characterization and diagnosis of disease. For instance, FISH was used to show that acute myelogenous leukemia (AML) is associated with a chromosome 16 inversion event near the centromere that leads to the fusion of two chromosome 16 genes: CBFB and MYH11 (Liu et al., 1993). FISH analyses have also contributed greatly to our understanding of Angelman syndrome and Prader-Willi syndrome (Knoll et al., 1989). Researchers found that Angelman syndrome and Prader-Willi syndrome were both associated with the same deletion in chromosome 15 (from region q11 to q13). However, they found that Angelman syndrome patients inherited the deleted copy of chromosome 15 from their mother, whereas Prader-Willi syndrome patients inherited the deleted copy of chromosome 15 from their father; this is due to the phenomenon known as chromosomal imprinting.
FISH can also be used to identify genes with increased copy number or to detect gene loss, as shown by more or fewer than two fluorescent "dots" in a somatic cell, respectively. Furthermore, FISH can be carried out using nondividing cells, which allows investigators to examine nonmitotic cells. This is important, because DNA packing is approximately 10,000 times less compact in nonmitotic (interphase) cells, allowing researchers to achieve a higher level of resolution. For example, the neurological disorder Charcot-Marie-Tooth type 1A is associated with a duplication of one million base pairs that can be resolved by interphase FISH, but not by metaphase FISH (Lupski et al., 1991).
An extremely high-resolution form of FISH, called fiber-FISH, is carried out using isolated chromosomes that are free from nuclear architecture and exist as long, stretched-out DNA fibers (Parra & Windle, 1993; Wiegant et al., 1992). By using DNA fibers as a template for FISH, researchers can resolve gene rearrangements and duplications with incredible precision.
Spectral Karyotyping (SKY) and Multiplex-FISH (M-FISH)
The ability to isolate individual human chromosomes using flow cytometry, combined with knowledge of the human genome sequence, has allowed cytogeneticists to develop 24-color probe sets that are used to label each human chromosome with a distinct color (Figure 3) . Chromosome-specific probes are made by labeling DNA fragments covering the length of each individual chromosome with a distinctly colored fluorescent dye, as shown in Figure 3a. The labeled DNA probes are then pooled and used in hybridization experiments with metaphase chromosome spreads. The labeled DNA probe sets bind to their complementary chromosomes, allowing each individual chromosome to be labeled with a specific fluorescent color along its entire length. In a somatic cell, the maternal and paternal copy of each chromosome will be labeled with the same colors, as shown in Figure 3b. This powerful approach, which permits the simultaneous tracking of all human chromosomes, has been called spectral karyotyping (SKY) or multiplex-FISH (M-FISH) (Schrock et al., 1996; Speicher et al., 1996).
These techniques are probably the most significant development in molecular cytogenetics in the past decade. Because each chromosome has its own color, chromosomal translocations are easily detected when a chromosome shows a region with a different color; moreover, the second color reports the identity of the other chromosome involved in the translocation. SKY/M-FISH techniques have allowed researchers to detect small chromosomal rearrangements in individuals with seemingly normal karyotypes and to determine more precisely the cytogenetic aberrations in individuals with complex aberrant karyotypes.
Comparative Genomic Hybridization
Next on the horizon for cytogeneticists was the ability to perform genome-wide scans to identify chromosomal regions associated with loss or gain of genetic information. In order to address this need, researchers developed a technique called comparative genome hybridization (CGH) (Figure 4). This approach involves the isolation and fragmentation of genomic DNA from both a control subject and an experimental subject. The fragmented control DNA sample is labeled with green fluorescence, and the fragmented experimental DNA sample is labeled with red fluorescence. The two DNA samples are pooled and used together as DNA probes in hybridization experiments with normal chromosomes. The green and red probes then compete to bind to the chromosomes.
For unaltered chromosomal regions, the green and red probes should bind equally, resulting in an orange/yellow color. However, if a chromosomal region was deleted in the experimental group, that region will appear more green under the microscope. Similarly, if a chromosomal region was amplified in the experimental group, the corresponding chromosomal region will appear more red under the microscope. With these patterns in mind, researchers can then scan along the length of the chromosomes to identify genomic alterations. Using this approach, researchers have discovered that the gene encoding the catalytic subunit of phosphatidylinositol 3-kinase (PIK3CA) is amplified in ovarian cancer (Shayesteh et al., 1999), thus identifying PIK3CA as an oncogene associated with ovarian cancer.
Standard CGH methods are very labor intensive and require the use of metaphase chromosomes, which leads to limited resolution. However, a more recent microarray-based CGH method does not require the use of metaphase chromosomes (Pinkel et al., 1998). This method uses arrays containing thousands of base-pair fragments of the human genome adhered to a microchip. Each individual DNA fragment, which is located in a specific position on the chip, corresponds to a known DNA sequence that has been mapped to a specific chromosomal region. The same color-coded probes (green for the control group, and red for the experimental group) are then used in hybridization experiments with the CGH microarray platform, which can be scanned using an automated approach. As described for standard CGH experiments, unaltered chromosome regions show equal binding of the green and red probes and a resulting orange/yellow color, whereas amplified and deleted chromosomal regions in the experimental group appear red and green, respectively. By using microarrays, researchers can very precisely determine the chromosomal regions and genes that are amplified or missing. In fact, the information derived from a single array CGH experiment is equal to that derived from thousands of FISH experiments.
Conclusion
The evolution of cytogenetic techniques and the mapping of the human genome have provided scientists with a great deal of insight into the causes of numerous genetic disorders. Though rooted in early chromosome staining and gene mapping techniques, modern FISH, SKY, and CGH methods have far outshone their predecessors by providing an unprecedented view of human chromosomes. These procedures allow for precise chromosome labeling and the identification of missing or translocated chromosomes or genes, and they have lessened cytogeneticists' reliance on the light microscope in favor of more high-tech devices. In the decades to come, cytogenetic technologies will be further refined, and the future of molecular cytogenetics looks bright. The field will most certainly continue to uncover new, unexpected insights about both human genetic structure and genetic disease.
References and Recommended Reading
Carrano, A. V., et al. Measurement and purification of human chromosomes by flow cytometry and sorting. Proceedings of the National Academy of Sciences 76, 1382–1384 (1979)
Donahue, R. P., et al. Probable assignment of the Duffy blood group locus to chromosome 1 in man. Proceedings of the National Academy of Sciences 61, 949–955 (1968)
Drets, M. E., & Shaw, M. W. Specific banding patterns of human chromosomes. Proceedings of the National Academy of Sciences 68, 2073–2077 (1971)
Druker, B. J. Perspectives on the development of a molecularly targeted agent. Cancer Cell 1, 31–36 (2002)
Ephrussi, B., & Weiss, M. C. Interspecific hybridization of somatic cells. Proceedings of the National Academy of Sciences 53, 1040–1042 (1965)
Ford, C. E., et al. A sex-chromosome anomaly in a case of gonadal dysgenesis (Turner's syndrome). Lancet 1, 711–713 (1959)
Harris, H., & Watkins, J. F. Hybrid cells derived from mouse and man: Artificial heterokaryons of mammalian cells from different species. Nature 205, 640–646 (1965) (link to article)
Heisterkamp, N., et al. Structural organization of the bcr gene and its role in the Ph' translocation. Nature 315, 758–761 (1985) (link to article)
Jacobs, P. A., & Strong, J. A. A case of human intersexuality having a possible XXY sex-determining mechanism. Nature 183, 302–303 (1959) (link to article)
Knoll, J. H., et al. Angelman and Prader-Willi syndromes share a common chromosome 15 deletion but differ in parental origin of the deletion. American Journal of Medical Genetics 32, 285–290 (1989)
Knudson, A. G., Jr. Mutation and cancer: Statistical study of retinoblastoma. Proceedings of the National Academy of Sciences 68, 820–823 (1971)
Landegent, J. E., et al. Chromosomal localization of a unique gene by non-autoradiographic in situ hybridization. Nature 317, 175–177 (1985) (link to article)
Lejeune, J., et al. Etude des chromosomes somatiques de neuf enfants mongoliens. [Study of somatic chromosomes from nine mongoloid children.] Comptes Rendus Hebdomadaires des Seances de l'Academie des Sciences 248, 1721–1722 (1959)
———. Three cases of partial deletion of the short arm of a 5 chromosome. Comptes Rendus Hebdomadaires des Seances de l'Academie des Sciences 257, 3098–3102 (1963)
Lele, K. P., et al. Chromosome deletion in a case of retinoblastoma. Annals of Human Genetics 27, 171–174 (1963)
Liu, P., et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science 261, 1041–1044 (1993)
Lupski, J. R., et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell 66, 219–232 (1991)
Nowell, P. C., & Hungerford, D. A. A minute chromosome in human chronic granulocytic leukemia. Science 142, 1497 (1960)
Parra, I., & Windle, B. High resolution visual mapping of stretched DNA by fluorescent hybridization. Nature Genetics 5, 17–21 (1993) doi:10.1038/ng0993-17 (link to article)
Pinkel, D., et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature Genetics 20, 207–211 (1998) doi:10.1038/2524 (link to article)
Rowley, J. D. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and Giemsa staining. Nature 243, 290–293 (1973) doi:10.1038/243290a0 (link to article)
Schrock, E., et al. Multicolor spectral karyotyping of human chromosomes. Science 273, 494–497 (1996)
Shayesteh, L., et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nature Genetics 21, 99–102 (1999) doi:10.1038/5042 (link to article)
Speicher, M. R., & Carter, N. P. The new cytogenetics: Blurring the boundaries with molecular biology. Nature Reviews Genetics 6, 782–792 (2005) doi:10.1038/nrg1692 (link to article)
Speicher, M. R., et al. Karyotyping human chromosomes by combinatorial multi-fluor FISH. Nature Genetics 12, 368–375 (1996) doi:10.1038/ng0496-368 (link to article)
Tjio, J. G., & Levan, A. The chromosome numbers of man. Hereditas 42, 1–6 (1956)
Trask, B. J. Human cytogenetics: 46 chromosomes, 46 years and counting. Nature Reviews Genetics 3, 769–778 (2002) doi:10.1038/nrg905 (link to article)
Wiegant, J., et al. High-resolution in situ hybridization using DNA halo preparations. Human Molecular Genetics 1, 587–591 (1992)




 Figure 1
Figure 1