« Prev Next »
What if you had a dresser drawer filled with thousands of puzzle pieces mixed together from an unknown number of jigsaw puzzles, and you wanted to solve each puzzle separately? How would you begin to sort this out? This puzzle drawer is analogous to what many researchers face when trying to discover the underlying genetic and environmental causes linked to complex forms of human disease.
What Is a Complex Disease?
For the most part, complex diseases are caused by a combination of genetic, environmental, and lifestyle factors, most of which have not yet been identified. The vast majority of diseases fall into this category, including several congenital defects and a number of adult-onset diseases. Some examples include Alzheimer's disease, scleroderma, asthma, Parkinson's disease, multiple sclerosis, osteoporosis, connective tissue diseases, kidney diseases, autoimmune diseases, and many more (Hunter, 2005).
Scientists now know that complex diseases do not obey the standard Mendelian patterns of inheritance. Although we inherit genes associated with these diseases, genetic factors represent only part of the risk associated with complex disease phenotypes. A genetic predisposition means that an individual has a genetic susceptibility to developing a certain disease, but this does not mean that a person harboring a genetic tendency is destined to develop the disease. The actual development of the disease phenotype depends in large part on a person's environment and lifestyle. While we cannot change our genes, we can alter our lifestyle and environment to prevent or delay the onset of such a disorder. Indeed, the interplay between genetic and environmental factors in complex disease continues to challenge researchers (Dempfle et al., 2008).
Considering gene-environment interactions can improve our understanding of the causes of complex disease, and it can also assist researchers in developing targeted therapies. Scientists now understand that gene products and the by-products of environmental insult often interact at the molecular level. Rather than studying genetic and environmental factors separately, researchers are now studying how genes and environmental factors interact with one another. By looking at the whole picture, researchers can identify genetic risk factors, which may in turn be modified in an environment-specific manner (Dempfle et al., 2008).
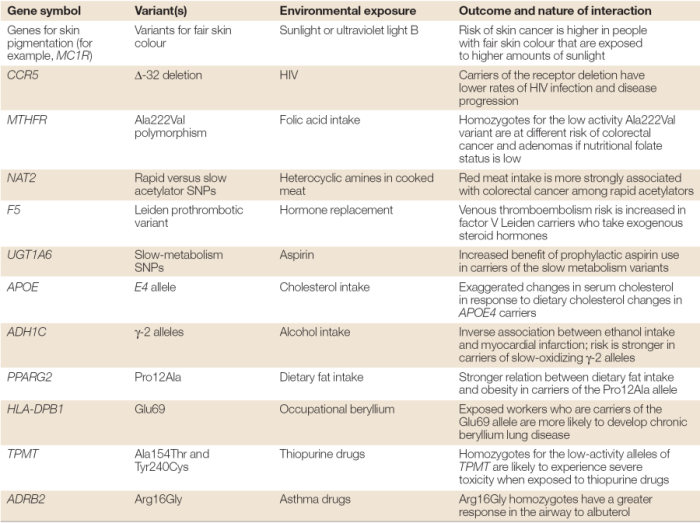
Because many studies of complex diseases have not examined the interplay among underlying genetic factors, there are few examples of reproducible data describing gene-environment interactions that influence disease (Wong et al., 2004). However, one good example of the interaction between genetic predisposition and environment is the higher susceptibility of fair-skinned people to the development of skin cancer. In these lighter-skinned individuals, it seems that skin cancer is associated with mutations in the melanocortin 1 receptor gene (MC1R). Another example of a genetic link to an environmental factor is seen among people with alcohol dehydrogenase (ADH) deficiency when they ingest even small amounts of alcohol. ADH deficiency is caused by a mutation in the ADH1C gene, which encodes an ADH subunit; mutations in the ADH1C gene lead to acute sensitivity to alcohol ingestion, common in individuals of Asian descent, and resulting in flushing of the face (Hunter, 2005). Some additional examples are shown in Table 1.
Returning to the jigsaw puzzle analogy, imagine that each puzzle piece has the ability to fit with more than one other puzzle piece, which could lead to more than one type of final "picture" when a puzzle is completed. Such is the prospect of disentangling the systems that regulate complex diseases, because we often do not understand the exact sense in which a complex disease is "complex." To sort out the causes of these conditions, researchers must dump the puzzle pieces out of the drawer and begin to assemble the jigsaw puzzle one piece at a time. The trick to solving the disease puzzle is to understand which factor—genetics or environment—plays a greater role in determining the course of a disease.
Understanding the Causes of Complex Disease
In general, a complex disease is one with features that complicate the detection of the disease's contributing factors. Discovering a contributing factor and characterizing its contribution to a complex disease is a difficult undertaking, because the effect of any single factor may be obscured or confounded by other contributing factors. Furthermore, genes and environment are both intangible variables.
To comprehend the intricacies of complex diseases, one must first understand Mendel's two main principles of inheritance: the principle of segregation and the principle of independent assortment. These principles explain how inherited traits, including those underlying disease, are passed from generation to generation. Since the mid-1800s, however, researchers have discovered many scenarios that violate Mendel's rules of inheritance, which can make determining the factors that influence complex genetic diseases difficult (Hern & Bidichandani, 2004). These phenomena include reduced penetrance, variable expressivity, phenotype definition (in cases in which a disease may be caused by more than one gene), polygenic traits, gene-gene interactions, and gene-environment interactions.
Complex diseases do not obey the single-gene dominant or single-gene recessive Mendelian pattern of inheritance (Davey & Ebrahim, 2003). In fact, diseases that exhibit simple Mendelian patterns of inheritance tend to be rare. Rather, complex diseases arise from numerous genetic and environmental factors working together. Despite this difference, studies of single-gene diseases can provide useful insights into the contribution of individual genes to the phenotypes associated with various complex diseases or conditions. A good example is Van der Woude syndrome, a disease caused by mutations in the IRF6 gene; this syndrome is associated with cleft lip and palate, lower lip pits, and occasional hypodontia (Kondo et al., 2002). Researchers have also discovered that IRF6 mutations are associated with isolated forms of cleft lip and palate (Zucchero et al., 2004), as well as isolated forms of hypodontia (Vieira et al., 2007). Both isolated cleft lip and palate and isolated hypodontia are multifactorial (complex) traits and much more common in the general population than Van der Woude syndrome.
The Human Genome Project and New Approaches in Gene Searching
Genomics is the field of study that seeks to understand the role of the genome in the development, function, and organization of cell types into higher order structures. The completion of the Human Genome Project has changed how researchers approach complex diseases by revealing new insights as to the genetic pathogenesis of disease. For example, scientists have discovered that the human genome sequence possesses numerous variations, of which single nucleotide polymorphisms (SNPs) are the most prevalent. As their name implies, SNPs are single DNA base-pair changes, located throughout our genome, that frequently vary from one person to another.
Scientists study the locations of SNPs to assess disease risk, and they also use these markers as a tool for the identification of disease-associated mutations. In particular, if a SNP occurs in a coding sequence of a gene that is involved in conferring disease susceptibility and the SNP disrupts the production of a functional gene product, then there is a high probability that this SNP will demonstrate a phenotypic effect. Also, scientists have constructed maps of SNPs, determining their locations along the length of every human chromosome. With these maps, studies can be designed to use SNPs as genetic markers. If a SNP appears to be segregating with a disease, or if it is more prevalent in affected versus unaffected subjects, this may indicate that the SNP is physically close to the disease-causing mutation (Cai et al., 2004). SNPs can also be used to delineate haplotypes, which are sets of closely linked alleles (genes or DNA polymorphisms) that are inherited together as a unit (Weiss & Terwilliger, 2000). Specifically, "tag" SNPs within haplotypes are identified and then used to uniquely identify those haplotypes.
The International HapMap Project is a collaborative effort to generate a catalog of SNP variants that occur in humans. It describes what these variants are, where they occur in the human DNA, and how they are distributed among people within populations and among populations in different parts of the world.
Evaluating Gene-Environment Interactions and Calculating Risk
Many studies have been conducted on either the genetic determinants or the environmental factors underlying complex disease, depending on the background of the researchers who are involved. Today, scientists are increasingly moving toward cross-disciplinary studies, collecting data on all facets of various diseases. However, additional multidisciplinary efforts are required to better monitor and elucidate disease development. For now, researchers primarily rely upon a number of complicated quantitative and statistical models in an attempt to determine complex disease risk.
Quantitative Models of Complex Disease Risk
Scientists struggle to define the risk patterns underlying complex diseases. This is because there are numerous combinations for modeling potential outcomes, even for the simplest of two-genotype scenarios (Wong et al., 2004; Hunter, 2005). These models strive to describe interactions between genetic susceptibility and environmental exposure for different diseases, such as ultraviolet light exposure for individuals with xeroderma pigmentosum (XP), a diet high in phenylalanine for individuals with phenylketonuria (PKU), and smoking for individuals with emphysema who carry a mutation in the AA1 gene. Disease risk is much higher when an individual with an at-risk gene variant is exposed to the disease-stimulating environment. Table 2 shows the complexity involved when attempting to calculate and model disease risk.
Table 2: Evaluating Disease Risk with Environmental and Genetic Contributions
| Gene Variant | Environmental Exposure | Relative Risk (XP) | Relative Risk (PKU) | Relative Risk (emphysema) |
| Absent | Absent | 1.0 | 1.0 | 1.0 |
| Present | Absent | ~1.0 | 1.0 | Modest |
| Absent | Present | Modest | 1.0 | Modest |
| Present | Present | Very High | Very High | High |
*Table repurposed from Hunter (2005)
Statistical Models of Complex Disease Risk
Scientists also use a variety of statistical modeling approaches to explain how genes and the environment contribute to specific disease phenotypes (Wong et al., 2004). For instance, Figure 1 depicts two models of gene-environment interactions that represent documented cases. Figure 1a considers individuals with two levels of susceptibility (genetically predisposed and not genetically predisposed) and two environmental exposure possibilities (exposed or non-exposed). This figure shows that the risk of disease development is greatest when a person has both genetic predisposition for the condition and environmental exposure. Figure 1b is similar, but it depicts a more complex scenario with three levels of environmental exposure. In this illustration, the risk of disease is elevated by moderate exposure to the environmental stimulus, yet it is not enhanced by exposure to a greater amount of the stimulus.
Both of these models are low in complexity when compared to a real-life complex disease state, but they do serve to illustrate the challenge faced by scientists and statisticians who are trying to understand underlying mechanisms associated with complex disease. Additionally, they illustrate how truly complex a disease state can be when it involves the potential overlap of multiple genetic factors, a variety of environmental factors, and different extents of environmental exposure (concentration, length of exposure, etc.).
Statistical Challenges of Modeling Complex Disease Risk
Statistical power for modeling complex diseases requires large numbers of people (sample sizes) from which to estimate the prevalence of a disease in the population. However, many complex diseases are rare; thus, scientists must rely on case-controlled studies to provide much-needed assessment of gene-environment interaction analyses. Case-controlled studies compare a group of patients who harbor the disease in question with a nonaffected patient group to identify factors that may contribute to the disease. These studies are less powerful than large-scale trials, but they are also less expensive and have led to many important medical discoveries (Schadt & Lum, 2006).
Another strategy being used to boost statistical power is "pooling" samples, or combining patient data from numerous studies. To this end, several consortia have formed to examine gene-environment interactions in prevalent forms of complex disease. For example, the U.S. National Cancer Institute (NCI) Breast and Prostate Cancer and Hormone-Related Gene Variants Cohort Consortium is examining more than 6,000 cases of breast cancer and 8,000 cases of prostate cancer, pooled across 10 prospective studies. To emphasize the potential power of this approach, the data from these consortia represent over 800,000 people who are being followed and a collective 7 million years of life (Hunter, 2005).
Genotype Dictates Response to Infectious Disease
Genes also dictate our susceptibility to some infectious diseases, as well as how well we respond to antiviral treatments. For example, our immune cells produce a protein called CCR5, which sits on the surface of our immune cells and serves as the gateway for the human immunodeficiency virus (HIV) to enter cells. Some people have mutations in their CCR5 gene that make them naturally immune to HIV infection, because they lack the CCR5 gateway that allows the virus into their cells. This fact suggested a novel way to prevent HIV infection—by developing drugs that block CCR5 function.
GWAS Help Unravel Complex Traits
According to the National Institutes of Health (NIH), genome-wide association studies (GWAS) encompass any study of genetic variation across the entire human genome that is designed to identify genetic associations with observable traits or the presence or absence of a disease or condition. GWAS are major tools for analyzing complex diseases, as they provide glimpses of the molecular pathways that lie beneath the disease landscape (Smyth et al., 2006).
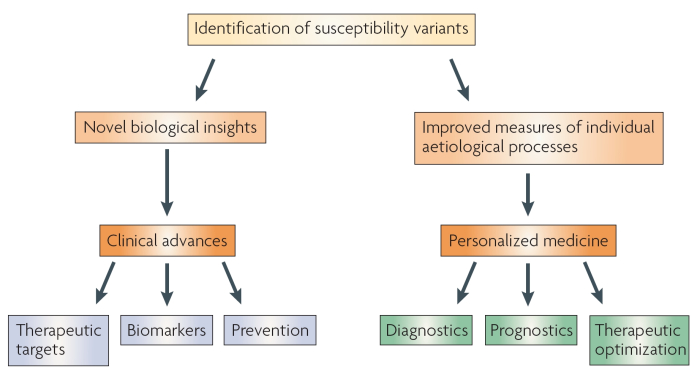
Some researchers argue that GWAS operate without a context for discerning what is being viewed, and hence they are of limited value (Chen & Witte, 2007). Regardless, they are being hailed as tools with the potential to facilitate personalized medicine and clinical care, as shown in the hierarchy presented in Figure 2. GWAS have provided novel insights and uncovered new sets of genes associated with a variety of human diseases, including obesity and breast cancer. The goal of GWAS is to translate these types of critical findings to clinical settings.
Personalized Medicine: Tailored Approaches to Preventing and Treating Complex Disorders
Hundreds of thousands of people die each year as a result of adverse drug reactions, which could be due to multiple factors, including disease determinants, environmental exposure, and genetic factors. Pharmacogenetics is the study of single-gene drug interactions, and pharmacogenomics focuses on many genes and their interactions with drugs. Researchers in these fields are undertaking studies on the genetic personalization of drug responses in an effort termed "personalized medicine" (Shastry, 2006). Because drug responses may be genotype-driven, the relationship between genotype and drug response may have a very real diagnostic value. Specifically, scientists are searching for biomarkers that can predict which patients will respond positively, which patients will be nonresponsive, and which patients will experience adverse reactions when prescribed the same medication and dose. Such personalized approaches to treatment hold great promise for improved health outcomes, including those related to complex disease.
References and Recommended Reading
Antonarakis, S. E., & Beckmann, J. S. Mendelian disorders deserve more attention. Nature Reviews Genetics 7, 277–282 (2006) doi:10.1038/nrg1826 (link to article)
Badano, J. L., & Katsanis, N. Beyond Mendel: An evolving view of human genetic disease transmission. Nature Reviews Genetics 3, 779–789 (2002) doi:10.1038/nrg910 (link to article)
Hunter, D. J. Gene–environment interactions in human diseases. Nature Reviews Genetics 6, 287–298 (2005) doi:10.1038/nrg1578 (link to article)
Guggino, W .B., & Stanton, B. A. New insights into cystic fibrosis: Molecular switches that regulate CFTR. Nature Reviews Molecular Cell Biology 7, 426–436 (2006) doi:10.1038/nrm1949 (link to article)
Jervis, G. A. Studies on phenylpyruvic oligophrenia: The position of the metabolic error. Journal of Biological Chemistry 169, 651–656 (1947)
———. Phenylpyruvic oligophrenia deficiency of phenylalanine-oxidizing system. Proceedings of the Society for Experimental Biology and Medicine 82, 514–515 (1953)
Nadeau, J. H. Modifier genes in mice and humans. Nature Reviews Genetics 2, 165–174 (2001) doi:10.1038/35056009 (link to article)
Paul, D. A double-edged sword. Nature 405, 515 (2000) doi:10.1038/35014676 (link to article)



 Table 1: Selected examples of gene-environment interactions observed in at least two studies.
Table 1: Selected examples of gene-environment interactions observed in at least two studies.