« Prev Next »
Early Discoveries of RNA Structure
Although there are multiple types of RNA molecules, the basic structure of all RNA is similar. Each kind of RNA is a polymeric molecule made by stringing together individual ribonucleotides, always by adding the 5'-phosphate group of one nucleotide onto the 3'-hydroxyl group of the previous nucleotide. Like DNA, each RNA strand has the same basic structure, composed of nitrogenous bases covalently bound to a sugar-phosphate backbone (Figure 1). However, unlike DNA, RNA is usually a single-stranded molecule. Also, the sugar in RNA is ribose instead of deoxyribose (ribose contains one more hydroxyl group on the second carbon), which accounts for the molecule's name. RNA consists of four nitrogenous bases: adenine, cytosine, uracil, and guanine. Uracil is a pyrimidine that is structurally similar to the thymine, another pyrimidine that is found in DNA. Like thymine, uracil can base-pair with adenine (Figure 2).
Single-stranded RNA can also form many secondary structures in which a single RNA molecule folds over and forms hairpin loops, stabilized by intramolecular hydrogen bonds between complementary bases. Such base-pairing of RNA is critical for many RNA functions, such as the ability of tRNA to bind to the correct sequence of mRNA during translation (Figure 3).
Robert Holley, a chemist at Cornell University, was the first researcher to work out the structure of tRNA (Holley et al., 1965). This molecule turned out to be the elusive structure that Francis Crick proposed in his so-called "adapter hypothesis" of 1955—a structure that carried amino acids and arranged them in a certain order that corresponded to the sequence in the nucleic acid strand. In 1968, Holley was awarded the Nobel Prize in Physiology or Medicine together with Gobind Khorana, at the University of Wisconsin, and Marshall Nirenberg, at the National Institutes of Health. Nirenberg and Khorana devised the key experiments to decipher the genetic code—in other words, which sequences of three nucleotides (codons) in an mRNA molecule would code for which amino acids.
mRNA and Splicing
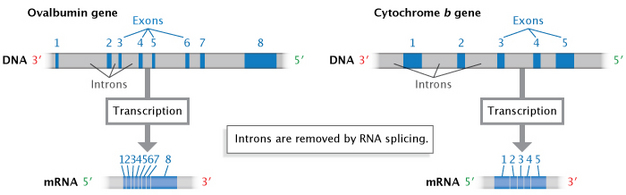
In eukaryotes (and to a lesser extent, prokaryotes), when RNA is first transcribed from DNA, it may contain additional noncoding sequences that are interspersed within the coding sequence. This immature RNA molecule is referred to as precursor mRNA (pre-mRNA) or heterogeneous nuclear RNA (hnRNA). The intervening noncoding sequences are called introns, and the segments of coding are known as material exons. The introns are then removed by a process known as RNA splicing to produce the mature mRNA molecule (Figure 7). An organelle called the spliceosome, composed of protein and small nuclear RNAs (snRNAs), is responsible for recognizing and removing the introns from pre-mRNA.
The surprising discovery of RNA splicing caused a paradigm shift in genetics. Much early work indicated that mRNA and the genes in DNA were colinear; that is, they were thought to match up, base for base, with the exception of the 3' poly(A) tail. In the late 1970s, however, seminal studies of gene expression in cells infected with an adenovirus demonstrated that the RNA transcripts produced by viral infection contained sequences that were not next to one another in the viral genome. Further study revealed that these mRNAs were produced after material had been removed or spliced out of a larger primary transcript (Berget et al., 1977; Evans et al., 1977). Since that time, introns have been found to occur in many eukaryotic cellular genes and some prokaryotic genes.
Probably the most thoroughly studied class of introns consists of those found in protein-coding genes. The 5' end of these introns almost always begins with the dinucleotide GU, and the 3' end typically contains AG. Changing one of these nucleotides precludes splicing. Another important sequence occurs at the branch point, anywhere from 18 to 40 nucleotides upstream from the 3' end of an intron. This sequence always contains an adenine, but it is otherwise loosely conserved. A typical sequence at a branch point is YNYYRAY, where Y indicates a pyrimidine, N denotes any nucleotide, R any purine, and A is for adenine (Figure 8) (Pierce, 2000; Patel & Steitz, 2003).
Many eukaryotic genes can be spliced in a number of different ways by choosing between different potential 5′ and 3′ splice junctions, thereby creating different combinations of exons and introns in the final mRNAs. This mix-and-match process allows the creation of several different proteins from a single gene sequence. The first example of such "alternative splicing" (Figure 9) was discovered in the adenovirus in 1977 (Berget et al., 1977). The first example in cellular genes was reported in 1980 in the IgM gene, which encodes an immunoglobulin, one of several proteins created by immune cells to fight infection by foreign organisms and particles (Early et al., 1980).
The Dscam gene of Drosophila, which encodes proteins involved in guiding embryonic nerves to their target destinations during formation of the fly's nervous system, exhibits an especially impressive number of alternative splicing patterns. Dozens of different forms of Dscam mRNAs and corresponding proteins have been identified, while analysis of the gene's sequence reveals a staggering 38,000 potential additional mRNAs, based on the large number of introns found. The ability to produce so many different proteins from a single gene may be necessary for forming as complex a structure as the nervous system (Schmucker et al., 2000). In general, the existence of multiple mRNA transcripts from single genes may account for the complexity of some organisms, such as humans, even though these organisms have relatively few genes (in the case of humans, approximately 25,000).
tRNA and rRNA: Their Role in Translation
Transfer RNA (tRNA) molecules serve as molecular adaptors that bind to mRNA on one end and carry amino acids into position on the other. Most types of cells possess approximately 30 to 40 different tRNAs, with more than one tRNA corresponding to each amino acid. tRNAs fold into a cloverleaf structure held together by the pairing of complementary nucleotides. Structural studies using X-ray crystallography have demonstrated that the cloverleaf is further folded into an L shape (Figure 10). A loop at one end of the folded structure base-pairs with three nucleotides on the mRNA that are collectively called a codon; the complementary three nucleotides on the tRNA are called the anticodon.
Although the pairing between codon and anticodon takes place over three nucleotides, strict complementary base-pairing is only necessary between the first two nucleotides. The third position is referred to as the "wobble" position (Figure 11), and the rules for base-pairing are less stringent at this position. Because of this flexibility, the 30 to 40 tRNAs present in a cell can "read" all 61 codons in mRNA.
The opposite end of the folded structure, which is the 3' end of the tRNA, binds to its corresponding amino acid at an attachment site that is also three nucleotides long, invariably CCA. Enzymes called aminoacyl-tRNA synthetases attach the correct amino acid to each tRNA, based on the three-dimensional structure of the tRNA molecule.
More and More RNAs
Other classes of RNA species include microRNAs, small interfering RNAs, and sRNAs—all of which are not translated into proteins but still perform important functions in the cell. The discovery of these RNAs has been one of the most exciting advances in recent years, and there is currently a lot of interest in the use of these molecules as possible therapies. But as far as their structure is concerned, these RNAs all share the same basic single-stranded chemical structure with, in some cases, higher-order structures obtained through complementary base-pair folding.
From the RNA Tie Club to today, the more scientists have studied RNA, the more surprises they have uncovered. New functions for RNA, new modifications to RNA, and other surprises undoubtedly await discovery in the years to come.

References and Recommended Reading
Berget, S. M., Moore, C., & Sharp, P. A. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proceedings of the National Academy of Sciences 74, 3171–3175 (1977)
Early, P., et al. Two mRNAs can be produced from a single immunoglobulin u chain by alternative RNS processing pathways. Cell 20, 313–319 (1980)
Evans, R. M., et al. The initiation sites for RNA transcription in Ad2 DNA. Cell 12, 733–739 (1977)
Holley, R. W., et al. Structure of a ribonucleic acid. Science 147, 1462–1465 (1965) doi:10.1126/science.147.3664.1462
Patel,
A. A., & Steitz, J. A. Splicing double: Insights from the second spliceosome.
Nature 4, 960–970 (2003) doi:10.1038/nrm1259 (link to article)
Pierce, B. A. Genetics: A Conceptual Approach, 2nd ed. (New York, Freeman, 2000)
Rich, A. A hybrid helix containing both deoxyribose and ribose polynucleotides and its relation to the transfer of information between the nucleic acids. Proceedings of the National Academy of Sciences 46, 1044–1053 (1960)
Rich, A., & Davies, D. R. A new two-stranded helical structure: Polyadenylic acid and polyuridylic acid.
Journal of the American Chemical Society
78, 3548–3549 (1956) (link to article)
Schmucker, D., et al. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101, 671–684 (2000)
Theimer, C. A., Blois, C. A., & Feigon, J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Molecular Cell 17, 671–682 (2005)



 Figure 1
Figure 1