« Prev Next »
The ERBB receptors are a group of receptor tyrosine kinases (RTKs) involved in key cellular functions, including cell growth and survival. They signal through phosphorylated tyrosines on their intracellular domains. Activation of these receptors leads to this phosphorylation and to subsequent intracellular signaling. What are the mechanisms of ERBB activation, and how did scientists discover them?
The Context for Transmembrane Signaling
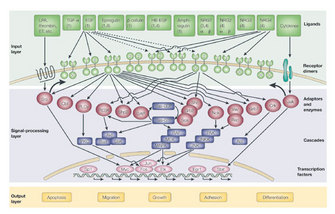
RTKs, a specific category of transmembrane receptors, can translate a ligand-binding event into a signal and can ultimately cause a wide array of cellular responses, including cell growth, differentiation, division, adhesion, migration, and apoptosis. One subfamily of RTKs contains the erythroblastic leukemia viral oncogene homologue (ERBB) receptors (Figure 1). This ERBB family of RTKs consists of four receptors: ERBB1, ERBB2, ERBB3, and ERBB4 (also known as HER1, HER2, HER3, and HER4). ERBB1 is also known as the epidermal growth factor (EGF) receptor.
How Can Receptor Activation Lead to Such a Wide Variety of Cellular Responses?
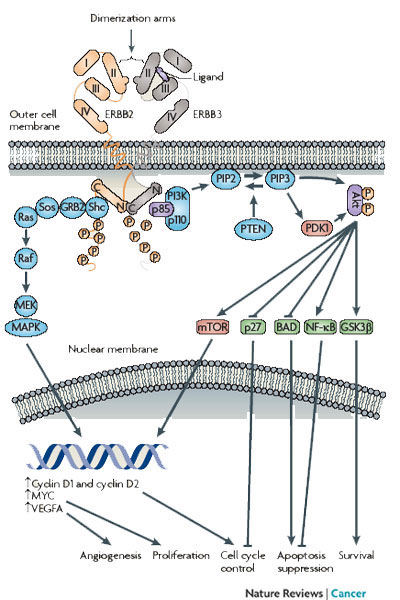
Each ERBB receptor has distinct locations and numbers of tyrosine phosphorylation sites on its C-terminal tails; there are twelve on ERBB1, six on ERBB2, and eleven on ERBB3 (Jones et al. 2006). ERBB4 also has phosphorylation sites, but they are less well characterized. Which ligand binds which receptor and what partner a receptor joins with (dimerization) affects which sites in the receptor become phosphorylated. Depending on which tyrosine residue of the two receptors in the dimer is phosphorylated, particular phosphotyrosine binding proteins will attach to the receptors. This attachment ultimately leads to propagation of the signal inside the cell and allows for multiple and varied cellular responses (Figure 2).
Growth Factor Ligands
ERBB receptors were discovered and characterized well after their ligands, soluble substances called growth factors. The earliest discovery of growth factors dates to the 1950s, when Rita Levi-Montalcini discovered a soluble substance called nerve growth factor (NGF) and Stanley Cohen discovered something quite similar, epidermal growth factor (EGF). The field of growth factor research began when Levi-Montalcini discovered that extracts from mouse tumor cells caused growth of neuronal cells in chicken embryos, and has since grown into a major area of scientific research concerning the growth, regeneration, and sustainment of biological tissues. In 1975, using radioactively labeled EGF and cells from human, mouse, rat, and chicken, Graham Carpenter and colleagues confirmed the presence of cell surface receptors that bind EGF, thereafter named EGF receptors, or EGFRs. Consequently, EGFRs were the first of the ERBB receptors for which the genetic sequence was defined and the size and function were determined.
What Forms Can ERBB Receptors Take?
ERBB receptors are monomeric, which means that they can exist as one receptor unattached to other receptors. These receptors can form homodimers (two of the same type of ERBB receptors bound together) and heterodimers (two different ERBB receptors bound together) in every possible combination. All the ERBB receptors except ERBB2 have known ligands, and all except ERBB3 have relatively active kinase domains. Although ERBB2 is minimally active in ligand binding, it is the preferred heterodimerization partner of the ERBB receptors. ERBB3 has shown minimal kinase activity, but this activity is much reduced compared with the other ERBB receptors. Interestingly, although both ERBB2 and ERBB3 have their own distinct deficiencies, together they form the most potent signaling pair in terms of mitogenic signaling (signals that cause cell division) (Table 1).
| Table 1: Kinase Activity and Ligand Binding Capability of ERBB Receptors | ||
|
Receptor
|
Kinase activity
|
Binds soluble ligand
|
| ERBB1 | Yes | Yes |
| ERBB2 | Yes | No |
| ERBB3 | Minimal | Yes |
| ERBB4 | Yes | Yes |
Ligand Binding and Receptor Phosphorylation
Ligand binding to the extracellular domain, which is the portion of the receptor on the outside of the cell, leads to several events. One is receptor dimerization or stabilization of preexisting inactive dimers (an intermolecular change), another is conformational changes in the receptors themselves (intramolecular change), and yet another is phosphorylation of C-terminal tyrosine residues. Often, in this context the term "activation" is equated with phosphorylation. When researchers are looking for activation of ERBB receptors in an experiment, they are usually looking for tyrosine phosphorylation quantity as a measure of the magnitude or occurrence of activation. They do so because when an RTK becomes activated, the result is phosphorylation of specific tyrosines on the cytoplasmic tail, the intracellular portion of the receptor. This phosphorylation leads to downstream signaling, which is the further modification of other molecules and proteins, either directly or indirectly, stemming from this phosphorylation of ERBB receptors.
Discovery of Dimer-Specific Phosphorylation
In 1989–1990, before any structural information about the kinase domain of the ERBB receptors was known, A. Honegger and colleagues published their observations about how EGFR becomes phosphorylated (Honegger et al. 1989, 1990). In their experiments, they asked if one EGFR could phosphorylate its dimerization partner. To find out, they constructed mutants that were uniquely deficient and expressed these mutant proteins in cells containing no endogenous EGFR. One mutant protein had a defective kinase domain, and one had a fully functional kinase domain but lacked its C-terminal tail. They found that the kinase-deficient EGFR indeed became phosphorylated, indicating that one receptor could phosphorylate its partner, and likely via an intramolecular interaction between the two receptors. These experiments provided evidence that receptors worked together to phosphorylate each other. But what occurs inside the kinase domain, and how exactly do the receptors interact to cause this phosphorylation?
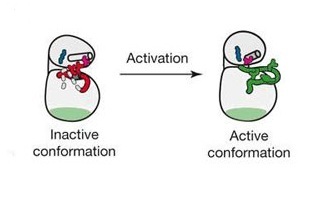
Defining ERBB Activation
To understand how activation of a receptor leads to phosphorylation of the cytoplasmic tail, let us first clarify what it means for a receptor to be activated. Like other RTKs, ERBB receptors can exist in an active or inactive state. Inside the heart of the kinase, the amino acid residues must be in certain places relative to one another for the kinase to function. A sequence of amino acids in the kinase domain, referred to as the activation loop, can move one way or the other, and that determines if the receptor is activated or not (Figure 3). What steps did scientists take to learn the basics about ERBB activation? EGFR has been the most extensively studied with respect to activation compared with the other ERBB receptors, so our discussion will focus EGFR, but much of the following is consistent for all the ERBB receptors.
What Is the Mechanism of ERBB Activation?
Until 2002, there were no structural data on the kinase domain of any of the ERBB receptors. Not until then was the structure of the kinase domain of EGFR solved, when researchers grew crystals containing their protein of interest, EGFR, and used a technique called X-ray crystallography to determine the three-dimensional structure of the protein, focusing on the business end: the kinase domain. Using this technique, J. Stamos and colleagues grew crystals containing EGFR and specifically obtained a crystal structure of the kinase domain. Analysis of this structure showed that the activation loop was in a position very close to the structurally similar insulin receptor when it is in an active state (Stamos, Sliwkowski & Eigenbrot 2002). In 2004, E. R. Wood and colleagues wanted to learn more about how a particular kinase inhibitor worked against EGFR, so they solved the structure of EGFR with this inhibitor (Lapatinib) bound to it (Wood et al. 2004). They found the structure of the kinase with the inhibitor bound to it, which was effectively the inactive state. At this point, it was still unclear which form of EGFR was the default conformation.
In a similar effort to find out the mechanisms of EGFR activation, X. Zhang and colleagues used crystallization conditions different from those used by previous groups: They used EGFR in a different solution. They also found EGFR in the active conformation and wondered why this conformation seemed to be favored when crystallized (Zhang et al. 2006). They found that by mutating a residue in the kinase domain, they were able to increase the activity of the kinase, which indicated that the kinase domain is normally under some form of inhibition. When they increased the concentration of the EGFR kinase domain in their experiment, they saw a concentration-dependent increase in kinase activity; that is, with high concentrations of kinase domain, they saw higher kinase activity. Intuitively, with higher kinase domain concentrations, each kinase domain would likely be closer to another and thus have a higher likelihood of activating one another. Would the converse also be true? Indeed, at lower concentrations, the researchers observed lower levels of kinase activity. These results are consistent with the possibility that the kinase domain is autoinhibited, or exists in a default state of inhibition. Why then would most of the crystallized forms of EGFR show the receptor in its active conformation? Most likely, the crystallization process necessary for analyzing protein structure caused a high concentration of receptors; thus, more of them were in the active state.
How Did Researchers Go About Learning about the Activation Mechanism?
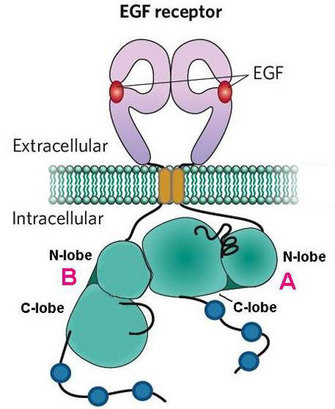
Some mechanisms of ERBB activation have been characterized, but there remain many open questions about this process. Some scientists suggest that dimerized kinase domains may switch places to activate one another (Zhang et al. 2006), but it is unclear if that actually happens, and if so, how they would do so. What triggers this switch? Furthermore, activation is regulated on many levels, both within the receptors themselves and through the proteins they associate with. We are still learning about the various mechanisms by which this process controls the magnitude and quenching of signaling.
Understanding the mechanisms by which receptors interact with one another and with other proteins emerges out of analyzing protein structures and perturbing the receptors' activated states. Through this work, activation itself can be better characterized. Activation of the ERBB receptors involves many components of the receptors, from the ligand binding in the extracellular domain to the kinase domain, and it ultimately leads to phosphorylation of tyrosines on the C-terminal tail. As a general principle, ligand binding leads to dimerization and subsequent activation of the receptors. Further, activation involves dimerization of the kinase domains in an asymmetric assembly. The structural features these receptors possess and the mechanisms they use to function continue to activate the curiosity of scientists.
References and Recommended Reading
Baselga, J. & Swain, S. M. Novel anticancer targets: Revisiting ERBB2 and discovering ERBB3. Nature Reviews Cancer 9, 463–475 (2009) doi:10.1038/nrc2656.
Burgess, A. W. et al. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Molecular Cell. 12, 541–552 (2003) doi:10.1016/S1097-2765(03)00350-2.
Citri, A. & Yarden, Y. EGF-ERBB signalling: Towards the systems level. Nature Reviews Molecular and Cellular Biology 7, 505–516 (2006) doi:10.1038/nrm1962.
Citri, A. et al. Hsp90 restrains ErbB-2/HER2 signalling by limiting heterodimer formation. EMBO reports 5, 1165–1170 (2004) doi:10.1038/sj.embor.7400300.
Groenen, L. C. et al. A model for the activation of the epidermal growth factor receptor kinase involvement of an asymmetric dimer? Biochemistry 36, 3826–3836 (1997) doi:10.1021/bi9614141.
Gschwind, A., Fischer, O. M. & Ullrich, A. The discovery of receptor tyrosine kinases: Targets for cancer therapy. Nature Reviews Cancer 4, 361–370 (2004) doi:10.1038/nrc1360.
Honegger, A. M. et al. Evidence that autophosphorylation of solubilized receptors for epidermal growth factor is mediated by intermolecular cross-phosphorylation. Proceedings of the National Academy of Sciences 86, 925–929.
Honegger, A. M. et al. Evidence for epidermal growth factor (EGF)-induced intermolecular autophosphorylation of the EGF receptors in living cells. Molecular and Cellular Biology 10, 4035–4044 (1990) doi:10.1016/S1097-2765(03)00350-2.
Jones, R. B. et al. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature 439, 168–174 (2006) doi:10.1038/nature04177.
Kuriyan, J. & Eisenberg, D. The origin of protein interactions and allostery in colocalization. Nature 450, 983–990 (2007) doi:10.1038/nature06524.
Shi, F. et al. ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proceedings of the National Academy of Sciences 107, 7692–7697 (2010) doi: 10.1073/pnas.1002753107.
Stamos, J., Sliwkowski, M. X. & Eigenbrot, C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. Journal of Biological Chemistry 277, 46265–46272 (2002) doi: 10.1074/jbc.M207135200.
Wood, E. R. et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): Relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Research 64, 6652–6659 (2004) doi:10.1158/0008-5472.
Yarden, Y. & Sliwkowski, M. X. Untangling the ErbB signalling network. Nature Reviews Molecular Cell Biology 2, 127–137 (2001) doi:10.1038/35052073.
Zhang X. et al. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125, 1137–1149 (2006) doi:10.1016/j.cell.2006.05.013.
Zhang, X. et al. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature 450, 741–744 (2007) doi:10.1038/nature05998.



 Figure 1: ERBB receptor signaling leads to a wide variety of signaling routes and outcomes
Figure 1: ERBB receptor signaling leads to a wide variety of signaling routes and outcomes


























