« Prev Next »

We live in an overcrowded world, and whether in search for food, shelter, or work, we have invaded new environments and disrupted ecosystems that have remained unchanged for thousands of years. As a result, many recently emerged pathogens are shared with wild animals. In particular, the ecosystem of the tropical forests is the most diverse in the world and it is harboring potentially threatening microbes. Non-human primates (NHP), our closest relatives, populate these ecosystems and constitute a reservoir of micro- and macro-parasites for humans, including a variety of helminths, protozoa, bacteria, and viruses. Over half of the shared pathogens listed as emerging in humans are viruses, and a large number of them have been isolated from wild NHPs (Jones et al. 2008). These include retroviruses like simian immunodeficiency virus, simian-T-lymphotrophic virus, and foamy virus (Nunn & Altizer 2006).
The zoonotic transmission of these pathogens from NHPs to humans may occur under different circumstances, including caring for captive NHPs, laboratory handling of tissues or fluids (Switzer et al. 2004), through exposure to sick or dead animals, or keeping NHPs as pets, with obvious consequences for human health (Wolfe et al. 1998). Similarly, the increased proximity to humans may put at greater risk already endangered or threatened populations of NHPs because of their susceptibility to human diseases like scabies, intestinal parasites, measles, and metapneumoviruses (Wallis & Lee 1999, Palacios et al. 2011). It is now widely accepted that a frequent mechanism of pathogen transmission from NHPs to humans is through the hunting and butchering of NHPs, because of the broad range of fluids and tissue types hunters and butchers come in contact with (Figure 1). These practices have been linked to the transmission of retroviruses such as adult T-cell leukaemia (HTLV-1) (Wolfe et al. 2005, Sintasath et al. 2009) and simian foamy virus (Calattini et al. 2007). However, the most striking and devastating example of an emerging disease resulting from cross-species transmission from NHPs is that of HIV/AIDS (Hahn et al. 2000). Since the recognition of the first cases in 1981 (Barre-Sinoussi et al. 1983), HIV/AIDS has infected over 33 million of people worldwide and has resulted in the death of 25 million of those infected (http://www.unaids.org/en/).

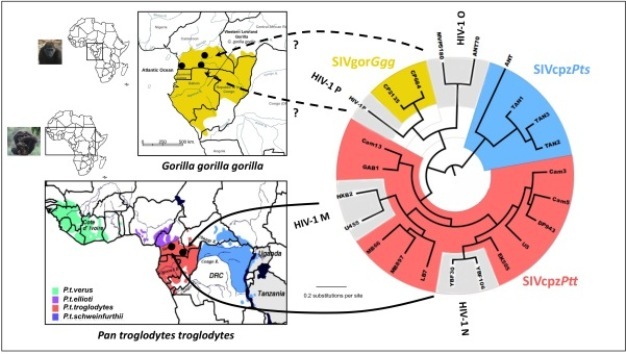
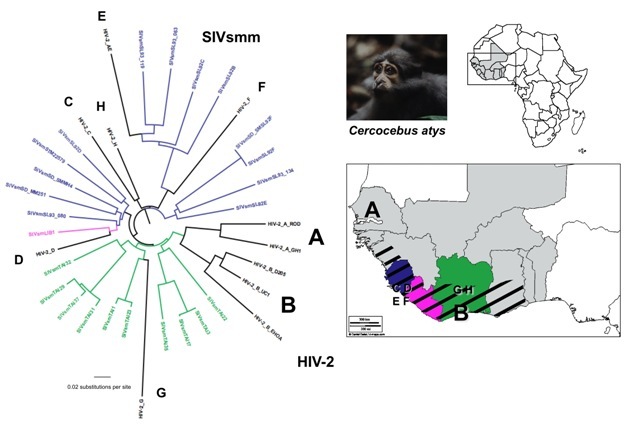
By comparing the gene sequences of human immunodeficiency viruses (HIV) and simian immunodeficiency viruses (SIV), virologists have identified different HIV groups, which correspond to 12 independent transmissions of SIV to humans. Pandemic HIV-1 group M, and group N descend from SIVcpz endemic in West Central African chimpanzees (Keele et al. 2006), while the closest relatives of HIV-1 groups O and P are SIVs infecting western lowland gorillas (SIVgor) in the same region (Van Heuverswyn et al. 2006, Plantier et al. 2009) (Figure 2). All known HIV-2 groups (A-H) descend from SIVsmm endemic in sooty mangabeys, which inhabit a strip of forested coast in West Africa (Hirsch et al. 1989, Gao et al. 1992) (Figure 3). Only HIV-1 M generated the global pandemic and 3 other strains were able to develop local epidemics in humans: HIV-1 group O in Cameroon, and HIV-2 groups A and B in West Africa (Hahn et al. 2000).
While HIV owes its origins in human populations to viral jumps from chimpanzees, gorillas, and sooty mangabeys, other NHPs living in Africa (such as Cercopithecus spp., Colobus spp., mandrills, drills, etc.) could also be candidates for potential zoonoses in the future, because they are heavily hunted for bushmeat and harbor their own SIVs (Aghokeng et al. 2010). There are 73 non-human primate species in Africa and more than 40 of these have tested positive for SIV so far (Table 1 and Figure 4). SIV prevalence varies depending on the species investigated, and when it is high, SIV can infect between 20% and 60% of the samples tested (Van de Woude & Apetrei 2006, Van Heuverswyn & Peeters 2007, Aghokeng et al. 2010). Interestingly, this virus is not found in Asian or South American NHPs, although no large surveys have yet been conducted on wild NHP species in these continents. This suggests that the last common ancestor of the catarrhines (Old World monkeys and apes) was not infected by SIV and therefore that SIV must have emerged after the radiation of these species in the last 25 million years (Sharp et al. 2000).
| Genus | Species Subspecies | Common name | SIV | References |
| Pan | troglodytes troglodytes | Central chimpanzee | SIVcpzPtt | (Corbet et al., 2000; Gao et al., 1999; Janssens et al., 1994; Peeters et al., 1989) |
|
troglodytes schweinfurthii |
East African chimpanzee |
SIVcpzPts |
(Peeters et al., 1992; Santiago et al., 2002) | |
|
Gorilla |
gorilla gorilla |
Western gorilla |
SIVgor |
(Van Heuverswyn et al., 2006) |
|
Colobus |
guereza |
Mantled guereza |
SIVcol |
(Courgnaud et al., 2001) |
|
Piliocolobus |
badius badius |
Western red colobus |
SIVwrcPbb |
(Courgnaud et al., 2003b; Liégeois et al., 2009) |
|
badius temminckii |
Temminck's Red Colobus |
SIVtrc |
(Locatelli et al., 2008a) | |
|
tholloni |
Thollon's Red Colobus |
SIVtrc |
(Ahuka-Mundeke et al., 2011) | |
|
rufomitratus |
Ugandan Red Colobus |
SIVtrc |
(Goldberg et al., 2009) | |
|
Procolobus |
verus |
Olive colobus |
SIVolc |
(Courgnaud et al., 2003b; Liégeois et al., 2009) |
|
Lophocebus |
aterrimus |
Black crested mangabey |
SIVbkm |
(Takemura et al., 2005) |
|
Papio |
cynocephalus |
Yellow baboon |
SIVagm-Ver |
(Jin et al., 1994b) |
|
ursinus |
Chacma baboon |
SIVagm-Ver |
(van Rensburg et al., 1998) | |
|
Cercocebus |
atys |
Sooty mangabey |
SIVsmm |
(Hirsch et al., 1989; Peeters et al., 1994) |
|
torquatus |
Red capped mangabey |
SIVrcm |
(Beer et al., 2001) | |
|
agilis |
Agile mangabey |
SIVagi |
(Nerrienet et al., 2002) | |
|
Mandrillus |
sphinx |
Mandrill |
SIVmnd-1,-2 |
(Souquiere et al., 2001; Tsujimoto et al., 1989) |
|
leucophaeus |
Drill |
SIVdrl |
(Clewley et al., 1998) | |
|
Miopithecus |
talapoin |
Angolan or southern talapoin |
SIVtal |
(Osterhaus et al., 1999) |
|
ogouensis |
Gabon or northern talapoin |
SIVtal |
(Liegeois et al., 2006) | |
|
Erythrocebus |
patas |
Patas monkey |
SIVagm-sab |
(Bibollet-Ruche et al., 1996) |
|
Chlorocebus |
sabaeus |
Green monkey |
SIVagm-Sab |
(Jin et al., 1994a) |
|
aethiops |
Grivet monkey |
SIVagm-Gri |
(Fomsgaard et al., 1991) | |
|
pygerithrus |
Vervet monkey |
SIVagm-Ver |
(Fukasawa et al., 1988) | |
|
tantalus |
Tantalus monkey |
SIVagm-Tan |
(Soares et al., 1997) | |
|
Cercopithecus |
nictitans |
Greater spot nosed monkey |
SIVgsn |
(Courgnaud et al., 2002) |
|
mitis |
Bleu monkey |
SIVblu |
(Bibollet-Ruche et al., 2000) | |
|
albogularis |
Sykes' monkey |
SIVsyk |
(Emau et al., 1991; Hirsch et al., 1993) | |
|
mona |
Mona monkey |
SIVmon |
(Barlow et al., 2003; Courgnaud et al., 2003a) | |
|
denti |
Dent's mona monkey |
SIVden |
(Dazza et al., 2005) | |
|
wolfi |
Wolf's mona monkey |
SIVwol |
(Ekwalanga et al., 2002; Saragosti et al., 2001) | |
|
cephus |
Mustached monkey |
SIVmus1,-2 |
(Aghokeng et al., 2007; Courgnaud et al., 2003a) | |
|
erythrotis |
Red-eared monkey |
SIVery |
(Worobey et al., 2010) | |
|
ascanius |
Red-tailed monkey |
SIVasc |
(Saragosti et al., 2001; Verschoor et al., 2004) | |
|
lhoest |
l'Hoest monkey |
SIVlho |
(Beer et al., 2000; Hirsch et al., 1999; Santiago et al., 2003b) | |
|
solatus |
Sun-tailed monkey |
SIVsun |
(Beer et al., 1999) | |
|
preussi |
Preuss's monkey |
SIVpre |
(Worobey et al., 2010) | |
|
neglectus |
De Brazza's monkey |
SIVdeb |
(Bibollet-Ruche et al., 2000) | |
|
Table 1: SIV infection in Old World monkeys and apes from Africa
The genus, species, and subspecies are given, as well as the common name. The corresponding SIV is described by a tree letter code (e.g., SIVdrl for drills). Only the non-human primate species in which SIVs have been confirmed by sequence analysis are listed here. Species representing a reservoir for HIV-1 and 2 are highlighted in bold. Species showing only serological evidence for SIV infection are not listed here. | ||||
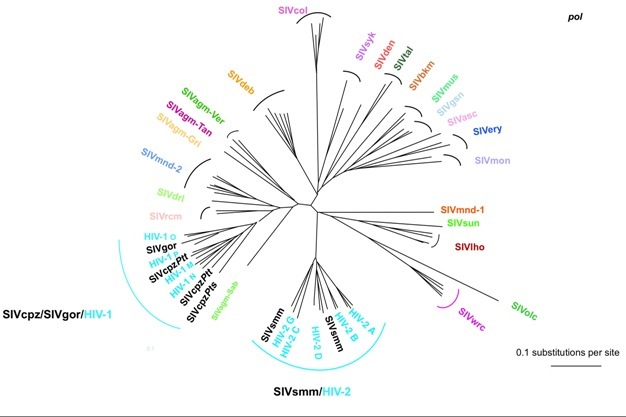
Figure 4: Evolutionary relationship among the different simian immunodeficiency viruses (SIV) and HIV lineages based on neighbor-joining phylogenetic analysis of partial sequences of the SIV gene polymerase (pol). The consensus length of the final alignment used for tree construction was 527 bp. Branch lengths are drawn to scale. Note that the different HIV-1 and HIV-2 lineages are interspersed with the SIVcpz/SIVgor and SIVsmm lineages respectively. Thus, from a phylogenetic point of view, the differentiation between HIVs and SIVs is irrelevant, which is the basis for the argument supporting the simian origins of HIV. The correspondence between the SIV lineages and their natural hosts are shown in Table 1.
Phylogenetic analyses of SIVs indicate a complex evolutionary history of association with their NHP hosts, reflecting thousands of years of host-virus co-speciation, cross-species transmission, superinfection, and recombination. Although in many instances SIVs are specific to the host species they infect, the SIV phylogenetic clusters are only partially superimposable on the primate phylogenetic trees. Some SIVs, such as SIVagm infecting four African monkeys species or SIVs of arboreal guenons, appear highly species-specific, with phylogenetic histories highly similar to those of their NHP hosts (Van de Woude & Apetrei 2006). However, there are also instances of viral host-switch, which could give the impression that a virus has been coevolving with its host, but in reality it is solely the result of a preferential cross-species transmission between genetically closely related host species (Wertheim & Worobey 2007). Well accepted examples of cross-species transmissions are those of SIVagm from African green monkeys, which has been transmitted t1998). The more scientists advanced in characterizing the SIV genomes, the more they realized that different parts of the SIV genome may be found in different speo Patas monkeys living in the same habitat in West Africa (Bibollet-Ruche et al. 1996), or to yellow and chacma baboons in South Africa (Jin et al. 1994b, van Rensburg et al. cies of non-human primates: several variants have discordant phylogenies when different genes are studied. These viruses have a mosaic genome structure: a cross-species transmission occurred followed by recombination of viruses of NHP which, at some point in time, shared the same habitat or ecological niches and got in direct contact with each other (through predation, habitat or food competition, or sexual contacts). For example, Mandrills are infected with two different viruses, SIVmnd-1 and SIVmnd-2, depending on whether they are living north or south of the Ogoué river in Gabon (Tsujimoto et al. 1989, Souquiere et al. 2001) and mustached monkeys are simultaneously infected with two very different viruses, SIVmus-1 and SIVmus-2 (Aghokeng et al. 2007). Because of recombination, which viruses represent ‘pure' lineages is still a topic of hot debate.
Expanding our understanding of the genetic diversity of SIVs in African primates and the monitoring of the virus' potential for recombination and transmission to new host species is particularly important because of the potential for new emerging infections in humans. For example, SIVcpz infecting chimpanzees of the troglodytes troglodytes subspecies, the precursor of the HIV-1 group M virus responsible for the AIDS pandemic, originated from the recombination of ancestors of SIVrcm infecting red-capped mangabeys and SIVgsn/mon/mus infecting greater spot-nosed monkeys, mustached monkeys, and mona monkeys. The recombination of these monkey viruses likely occurred within chimpanzees, as these apes are known to hunt other small primates for food (Bailes et al. 2003). As we now know, recombinant SIVcpz was again able to cross the species barrier into gorillas and humans and initiate one of the deadliest pandemics in human history.
The distribution of SIV may also provide information about the timing of its origin and evolutionary history, as well as potentially identifying populations that have evolved resistance to the virus. For example, SIVcpz infections appear absent in Pan troglodytes verus, a chimpanzee subspecies occupying the Upper Guinea region of West Africa (Prince et al. 2002, Santiago et al. 2002, Switzer et al. 2005). In addition, despite frequent contacts with SIVwrc through hunting of highly infected Western red colobus monkeys (prevalence of 50-80%, Locatelli et al. 2008b, Leendertz et al. 2010), the absence of SIVwrc-like sequences is also surprising in P. troglodytes verus (Leendertz et al. 2011) (Figure 5). Another subspecies, Pan troglodytes ellioti, inhabiting the Gulf of Guinea region north of the Sanaga river of Cameroon, is similarly SIV free (Van Heuverswyn et al. 2007). So far, only chimpanzee subspecies found south of the Sanaga river (Pan troglodytes troglodytes and Pan troglodytes schweinfurthii) have tested positive for SIVcpz (Santiago et al. 2003a, Van Heuverswyn et al. 2007). Scientists are currently examining four hypotheses to explain the apparent lack of SIV in chimpanzee subspecies north of the Sanaga river. First, the Sanaga may represent a barrier to both gene flow in chimpanzee subspecies and viral transmission. This would suggest an introduction of SIV into chimpanzees following subspecies divergence. A recent study by Gonder et al. (2011), estimated that chimpanzees living north of the Sanaga river shared a last common ancestor with chimpanzees from southern Cameroon 0.32 mya and these estimates are consistent with previously reported studies. Second, it is possible that SIVwrc is incapable of jumping between species in a way similar to SIVrcm and SIVgsn. SIVs vary in intrinsic properties enabling them to adapt to new hosts by overcoming cellular defenses. The inability of some viruses to adapt to new hosts may explain why more than 40 NHP species are infected by species-specific SIVs at relatively high prevalence, but only SIVs from sooty mangabeys, chimpanzees, or gorillas have been transmitted on multiple occasions to the human population. Third, it is possible that subspecies north of the Sanaga have historically been infected with SIV but have evolved resistance resulting in the local extinction of SIV. Lastly, it is possible, though unlikely, that P. t. verus and P. t. ellioti are infected by SIV strains that have so far eluded detection or that are at very low prevalence in these populations.

SIV infections in most NHPs appear avirulent, which has been taken as evidence of a prolonged coevolutionary history (Silvestri et al. 2007). However, heterologous infections do represent a threat, as confirmed by the consequences of cross-species transmissions of SIVcpz and SIVsmm into the human population and of SIVrcm and SIVgsn into SIVcpz. A recent study on habituated communities of wild Pan troglodytes schweinfurthii chimpanzees in Gombe, Tanzania, reported that SIVcpzPts infection has a similar effect on chimpanzees as HIV-1 has on humans (Keele et al. 2009). Similarly, a report on a naturally SIV infected P.t.troglodytes chimpanzee confiscated in Cameroon in 2003, suggests clinical progression to an AIDS-like disease in this animal (Etienne et al. 2011). Viral populations evolve faster than host populations, therefore recognition of a new virus by the host cell will always lag behind, unless coincidental viral restriction already exists in the newly infected host. This means that the restriction factors that each species currently possesses, mainly protect that species from past infections but not current ones (Sawyer et al. 2005, Gupta & Towers 2009).
If successful cross-species transmission does occur, there is still no guarantee that this will be followed by efficient spread into the new population. Other factors, such as transmission between hosts in the new species as well as other factors unique to the environment of the new host, need to be taken into account. This is illustrated by differences in geographic distribution and virulence of HIV-1, the pandemic virus, and HIV-2, an epidemic mostly confined in western Africa.
The opportunity for transmission of SIVs (or other pathogens) continues to increase as humans encroach on the natural environment. In West and Central Africa, hunting has shifted from a primarily subsistence activity to an organized, commercial business (Laporte et al. 2007). As a consequence, more individuals are potentially exposed to wild reservoirs of pathogens. Commercial logging in Equatorial Africa has led to road construction in remote forested areas; these roads have facilitated human migration into previously uninhabited regions and have precipitated changes in social and economic networks (including commercial sex work) that support this industry (Poulsen et al. 2009). Unfortunately, HIV prevalence is increasing in rural areas. The possibility of recombination between newly introduced SIVs and circulating HIVs can pose an additional risk for the outbreak of a novel epidemic and further threaten the survival of immunocompromised people exposed to new pathogens (Laurent et al. 2004, LeBreton et al. 2007).
Other retroviruses are also very telling and interesting from a host-virus dynamic and evolutionary perspective. The simian foamy viruses (SFVs) have frequently been transmitted to humans exposed to bushmeat, apparently without causing disease and further spread. The congruence in the phylogeny of SFVs and that of their NHP hosts is evidence of tight virus-host co-evolution occurring over the last 30-40 million years (Switzer et al. 2005, Liu et al. 2008). A very different picture emerges with epidemic human T-cell lymphotropic viruses (HTLV) associated with certain forms of leukemia, which entered the human population from their simian counterparts (STLV) (Wolfe et al. 2005). STLV transmissions between NHP species have been described, suggesting the ease with which STLVs cross species barriers (Liégeois et al. 2008). As a consequence, STLV infections are phylogenetically grouped according to geography rather than reflecting the phylogenetic history of the hosts. These differences in the incidence of between species transmission may have utility for scientists in other settings. For example, because of the tight co-evolution between SFV and their NHP hosts, the study of SFV may aid in resolving or clarifying the debate about the taxonomic classification of certain NHP species. Further study of STLVs may provide new information about the contact established between different NHP species.
For emerging infectious diseases of humans such as the AIDS pandemic, avian influenza, Ebola, and SARS, the question of their origin is extremely important. Equally important is the question of why certain animal viruses fail to launch sustained human-to-human transmissions. In conclusion, the current HIV-1 pandemic demonstrates that the transmission of NHP lentiviruses into the human population can have unexpected and very serious consequences. Furthermore, additional cross-species transmissions of NHP lentiviruses other than those of chimpanzee, gorilla, or sooty mangabey origin may already have happened, just waiting for a chance to become the next global plague.
References and Recommended Reading
Aghokeng, A. F. et al. Extensive survey on the prevalence and genetic diversity of SIVs in primate bushmeat provides insights into risks for potential new cross-species transmissions. Infection, Genetics and Evolution 10, 386-396 (2010).
Ahuka-Mundeke, S. et al. Novel multiplexed HIV/simian immunodeficiency virus antibody detection assay. Emerging Infectious Diseases 17, 2277-2286 (2011). Available from http://dx.doi.org/10.3201/eid1712.110783
Bailes, E. et al. Hybrid origin of SIV in chimpanzees. Science 300, 1713 (2003).
Barlow, K. L., Ajao, A. O. & Clewley, J. P. Characterization of a novel simian immunodeficiency virus (SIVmonNG1) genome sequence from a mona monkey (Cercopithecus mona). Journal of Virology 77, 6879-6888 (2003).
Barre-Sinoussi, F. et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220, 868-871 (1983).
Beer, B. E. et al. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): Evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. Journal of Virology 73, 7734-7744 (1999).
Beer, B. E. et al. Patterns of genomic sequence diversity among their simian immunodeficiency viruses suggest that L'Hoest monkeys (Cercopithecus lhoesti) are a natural lentivirus reservoir. Journal of Virology 74, 3892-3898 (2000).
Beer, B. E. et al. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). Journal of Virology 75, 12014-12027 (2001).
Bibollet-Ruche, F. et al. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): Evidence for cross-species transmission from African green monkeys (Cercopithecus aethiops sabaeus) in the wild. Journal of General Virology 77, 773-781 (1996).
Bibollet-Ruche, F. et al. Molecular characterization of new primate lentiviruses from De Brazza, mona, and blue monkeys: Evidence for host-dependent evolution within this group of viruses. Proceedings of the 7th Conference on Retroviruses and Opportunistic Infections, Abstract 223 (2000). (link)
Calattini, S. et al. Simian foamy virus transmission from apes to humans, rural Cameroon. Emerging Infectious Diseases 13, 1314-1320 (2007). Available from http://wwwnc.cdc.gov/eid/article/13/9/06-1162_article.htm
Clewley, J. P. et al. A novel simian immunodeficiency virus (SIVdrl) pol sequence from the drill monkey, Mandrillus leucophaeus. Journal of Virology 72, 10305-10309 (1998).
Corbet, S. et al. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. Journal of Virology 74, 529-534 (2000).
Courgnaud, V. et al. Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: A new lineage in the nonhuman primate lentivirus family. Journal of Virology 75, 857-866 (2001).
Courgnaud, V. et al. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. Journal of Virology 76, 8298-8309 (2002).
Courgnaud, V. et al. Identification of a new simian immunodeficiency virus lineage with a vpu gene present among different cercopithecus monkeys (C. mona, C. cephus, and C. nictitans) from Cameroon. Journal of Virology 77, 12523-12534. (2003a).
Courgnaud, V. et al. Partial molecular characterization of two simian immunodeficiency viruses (SIV) from African colobids: SIVwrc from Western red colobus (Piliocolobus badius) and SIVolc from olive colobus (Procolobus verus). Journal of Virology 77, 744-748 (2003b).
Dazza, M. C. et al. Characterization of a novel vpu-harboring simian immunodeficiency virus from a Dent's Mona monkey (Cercopithecus mona denti). Journal of Virology 79, 8560-8571 (2005).
Ekwalanga, M. et al. Molecular characterization of a primate lentivirus from Cercopithecus wolfi. XIV International AIDS Conference, Barcelona, Spain (2002). Available from http://www.aegis.org/conferences/iac/2002/TuPeA4417.html
Emau, P. et al. Isolation from African Sykes' monkeys (Cercopithecus mitis) of a lentivirus related to human and simian immunodeficiency viruses. Journal of Virology 65, 2135-2140 (1991).
Etienne, L. et al. Characterization of a new simian immunodeficiency virus strain in a naturally infected Pan troglodytes troglodytes chimpanzee with AIDS related symptoms. Retrovirology 8, 4 (2011). doi:10.1186/1742-4690-8-4
Fomsgaard, A. et al. A highly divergent proviral DNA clone of SIV from a distinct species of African green monkey. Virology 182, 397-402 (1991).
Fukasawa, M. et al. Sequence of simian immunodeficiency virus from African green monkey, a new member of the HIV/SIV group. Nature 333, 457-461 (1988).
Gao, F. et al. Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature 358, 495-499 (1992).
Gao, F. et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397, 436-441 (1999).
Goldberg, T. L. et al. Coinfection of Ugandan red colobus (Procolobus [Piliocolobus] rufomitratus tephrosceles) with novel, divergent delta-, lenti-, and spumaretroviruses. Journal of Virology 83, 11318-11329 (2009).
Gonder, M. K. et al. Evidence from Cameroon reveals differences in the genetic structure and histories of chimpanzee populations. Proceedings of the National Academy of Sciences of the United States of America 108, 4766-4771 (2011).
Gupta, R. K. & Towers, G. J. A tail of Tetherin: How pandemic HIV-1 conquered the world. Cell Host & Microbe 6, 393-395 (2009).
Hahn, B. H. et al. AIDS as a zoonosis: Scientific and public health implications. Science 287, 607-614 (2000).
Hirsch, V. M. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339, 389-392 (1989).
Hirsch, V. M. et al. A distinct African lentivirus from Sykes' monkeys. Journal of Virology 67, 1517-1528 (1993).
Hirsch, V. M. et al. Characterization of a novel simian immunodeficiency virus (SIV) from L'Hoest monkeys (Cercopithecus l'hoesti): Implications for the origins of SIVmnd and other primate lentiviruses. Journal of Virology 73, 1036-1045 (1999).
Janssens, W. et al. Phylogenetic analysis of a new chimpanzee lentivirus SIVcpz-gab2 from a wild-captured chimpanzee from Gabon. AIDS Research and Human Retroviruses 10, 1191-1192 (1994).
Jin, M. J. et al. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. The EMBO Journal 13, 2935-2947 (1994a).
Jin, M. J. et al. Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: Evidence for cross-species transmission in the wild. Journal of Virology 68, 8454-8460 (1994b).
Jones, K. E. et al. Global trends in emerging infectious diseases. Nature 451, 990-993 (2008).
Keele, B. F. et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313, 523-526 (2006).
Keele, B. F. et al. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature 460, 515-519 (2009).
Laporte, N. T. et al. Expansi wassssg and HIV epidemic, rural Equatorial Africa. Emerging Infectious Diseases 10, 1953-1956 (2004). Available from http://wwwnc.cdc.gov/eid/article/10/11/04-0180.htm
LeBreton, M. et al. Exposure to wild primates among HIV-infected persons. Emerging Infectious Diseases 13, 1579-1582 (2007). Available from http://wwwnc.cdc.gov/eid/article/13/10/07-0338.htm
Leendertz, S. A. et al. High prevalence, coinfection rate, and genetic diversity of retroviruses in wild red colobus monkeys (Piliocolobus badius badius) in Tai National Park, Cote d'Ivoire. Journal of Virology 84, 7427-7436 (2010).
Leendertz, S. A. et al. No evidence for transmission of SIVwrc from western red colobus monkeys (Piliocolobus badius badius) to wild West African chimpanzees (Pan troglodytes verus) despite high exposure through hunting. BMC Microbiology 11, 24 (2011). doi:10.1186/1471-2180-11-24
Liegeois, F. et al. Molecular characterization of a novel simian immunodeficiency virus lineage (SIVtal) from northern talapoins (Miopithecus ogouensis). Virology 349, 55-65 (2006).
Liégeois, F. et al. Identification and molecular characterization of new STLV-1 and STLV-3 strains in wild-caught nonhuman primates in Cameroon. Virology 371, 405-417 (2008).
Liégeois, F. et al. Full-length genome characterization of a novel Simian Immunodeficiency Virus lineage (SIVolc) from Olive Colobus (Procolobus verus) and new SIVwrcPbb strains from Western Red Colobus (Piliocolobus badius badius) from the Taï Forest in Ivory Coast. Journal of Virology 83, 428-439 (2009).
Liu, W. et al. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathogens 4, e1000097 (2008). doi:10.1371/journal.ppat.1000097
Locatelli, S. et al. Full molecular characterization of a simian immunodeficiency virus, SIVwrcpbt from Temminck's red colobus (Piliocolobus badius temminckii) from Abuko Nature Reserve, The Gambia. Virology 376, 90-100 (2008a).
Locatelli, S. et al. Prevalence and genetic diversity of simian immunodeficiency virus infection in wild-living red colobus monkeys (Piliocolobus badius badius) from the Tai forest, Cote d'Ivoire SIVwrc in wild-living western red colobus monkeys. Infection, Genetics and Evolution 8, 1-14 (2008b).
Nerrienet, E. et al. New primate lentivirus from agile monkeys (Cercocebus agilis) suggests a host-dependant evolution within mangabeys in Central Africa. XIV International AIDS Conference, Barcelona, Spain (2002). Available from http://www.iasociety.org/Abstracts/A7062.aspx
Nunn, C. L. & Altizer, S. Infectious Diseases in Primates. Behavior, Ecology and Evolution. Oxford, UK: Oxford University Press, 2006.
Osterhaus, A. D. et al. Isolation and partial characterization of a lentivirus from talapoin monkeys (Myopithecus talapoin). Virology 260, 116-124 (1999).
Palacios, G. et al. Human metapneumovirus infection in wild mountain gorillas, Rwanda. Emerging Infectious Diseases 17, (2011). Available from http://dx.doi.org/10.3201/eid1704.100883
Peeters, M. et al. Isolation and partial characterization of an HIV-related virus occurring naturally in chimpanzees in Gabon. AIDS 3, 625-630 (1989).
Peeters, M. et al. Isolation and characterization of a new chimpanzee lentivirus (simian immunodeficiency virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS 6, 447-451 (1992).
Peeters, M. et al. Isolation of simian immunodeficiency viruses from two sooty mangabeys in Cote d'Ivoire: Virological and genetic characterization and relationship to other HIV type 2 and SIVsm/mac strains. AIDS Research and Human Retroviruses 10, 1289-1294 (1994).
Plantier, J. C. et al. A new human immunodeficiency virus derived from gorillas. Nature Medicine 15, 871-872 (2009).
Poulsen, J. R. et al. Bushmeat supply and consumption in a tropical logging concession in northern Congo. Conservation Biology 23, 1597-1608 (2009).
Prince, A. M. et al. Lack of evidence for HIV type 1-related SIVcpz infection in captive and wild chimpanzees (Pan troglodytes verus) in West Africa. AIDS Research and Human Retroviruses 18, 657-660 (2002).
Santiago, M. L. et al. SIVcpz in wild chimpanzees. Science 295, 465 (2002).
Santiago, M. L. et al. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii). Journal of Virology 77, 7545-7562 (2003a).
Santiago, M. L. et al. Noninvasive detection of Simian immunodeficiency virus infection in a wild-living L'Hoest's monkey (Cercopithecus Ihoesti). AIDS Research and Human Retroviruses 19, 1163-1166 (2003b).
Saragosti, S. et al. Molecular characterization of primate lentiviruses from a Cercopithecus wolfi and a Cercopithecus ascanius. Abstract, p19, 8th Annual Discussion Meeting on HIV Dynamics and Evolution, Paris, France (2001).
Sawyer, S. L. et al. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proceedings of the National Academy of Sciences of the United States of America 102, 2832-2837 (2005).
Sharp, P. M. et al. Origins and evolution of AIDS viruses: Estimating the time-scale. Biochemical Society Transactions 28, 275-282 (2000).
Silvestri, G. et al. Understanding the benign nature of SIV infection in natural hosts. Journal of Clinical Investigation 117, 3148-3154 (2007).
Sintasath, D. M. et al. Simian T-lymphotropic virus diversity among nonhuman primates, Cameroon. Emerging Infectious Diseases 15, 175-184 (2009). Available from http://wwwnc.cdc.gov/eid/article/15/2/08-0584.htm
Soares, M. A. et al. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology 228, 394-399 (1997).
Souquiere, S. et al. Wild Mandrillus sphinx are carriers of two types of lentivirus. Journal of Virology 75, 7086-7096 (2001).
Switzer, W. M. et al. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. Journal of Virology 78, 2780-2789 (2004).
Switzer, W. M. et al. The epidemiology of simian immunodeficiency virus infection in a large number of wild- and captive-born chimpanzees: Evidence for a recent introduction following chimpanzee divergence. AIDS Research and Human Retroviruses 21, 335-342 (2005).
Switzer, W. M. et al. Ancient co-speciation of simian foamy viruses and primates. Nature 434, 376-380 (2005).
Takemura, T. et al. A novel simian immunodeficiency virus from black mangabey (Lophocebus aterrimus) in the Democratic Republic of Congo. Journal of General Virology 86, 1967-1971 (2005).
Tsujimoto, H. et al. Sequence of a novel simian immunodeficiency virus from a wild-caught African mandrill. Nature 341, 539-541 (1989).
Van de Woude, S. & Apetrei, C. Going wild: Lessons from naturally occurring T-lymphotropic lentiviruses. Clinical Microbiology Reviews 19, 728-762 (2006).
Van Heuverswyn, F. & Peeters, M. The origins of HIV and implications for the global epidemic. Current Infectious Disease Reports 9, 338-346 (2007).
Van Heuverswyn, F. et al. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444, 164 (2006).
Van Heuverswyn, F. et al. Genetic diversity and phylogeographic clustering of SIVcpzPtt in wild chimpanzees in Cameroon. Virology 368, 155-171 (2007).
van Rensburg, E. J. et al. Simian immunodeficiency viruses (SIVs) from eastern and southern Africa: Detection of a SIVagm variant from a chacma baboon. Journal of General Virology 79, 1809-1814 (1998).
Verschoor, E. J. et al. A novel simian immunodeficiency virus isolated from a Schmidt's guenon (Cercopithecus ascanius schmidti). Journal of General Virology 85, 21-24 (2004).
Wallis, J. & Lee, D. R. Primate conservation: The prevention of disease transmission. International Journal of Primatology 20, 803-826 (1999).
Wertheim, J. O. & Worobey, M. A challenge to the ancient origin of SIVagm based on African green monkey mitochondrial genomes. PLoS Pathogens 3, e95 (2007). doi:10.1371/journal.ppat.0030095
Wolfe, N. D. et al. Wild primate populations in emerging infectious disease research: The missing link? Emerging Infectious Diseases 4, 149-158 (1998). Available from http://wwwnc.cdc.gov/eid/article/4/2/98-0202.htm
Wolfe, N. D. et al. Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proceedings of the National Academy of Sciences of the United States of America 102, 7994-7999 (2005).
Worobey, M. et al. Island biogeography reveals the deep history of SIV. Science 329, 1487 (2010).