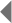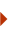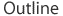DNA replication is a truly amazing biological phenomenon. Consider the countless number of times that your cells divide to make you who you are—not just during development, but even now, as a fully mature adult. Then consider that every time a human cell divides and its DNA replicates, it has to copy and transmit the exact same sequence of 3 billion nucleotides to its daughter cells. Finally, consider the fact that in life (literally), nothing is perfect. While most DNA replicates with fairly high fidelity, mistakes do happen, with polymerase enzymes sometimes inserting the wrong nucleotide or too many or too few nucleotides into a sequence. Fortunately, most of these mistakes are fixed through various DNA repair processes. Repair enzymes recognize structural imperfections between improperly paired nucleotides, cutting out the wrong ones and putting the right ones in their place. But some replication errors make it past these mechanisms, thus becoming permanent mutations. These altered nucleotide sequences can then be passed down from one cellular generation to the next, and if they occur in cells that give rise to gametes, they can even be transmitted to subsequent organismal generations. Moreover, when the genes for the DNA repair enzymes themselves become mutated, mistakes begin accumulating at a much higher rate. In eukaryotes, such mutations can lead to cancer.
Errors Are a Natural Part of DNA Replication
After James Watson and Francis Crick published their model of the double-helix structure of DNA in 1953, biologists initially speculated that most replication errors were caused by what are called tautomeric shifts. Both the purine and pyrimidine bases in DNA exist in different chemical forms, or tautomers, in which the protons occupy different positions in the molecule (Figure 1). The Watson-Crick model required that the nucleotide bases be in their more common "keto" form (Watson & Crick, 1953). Scientists believed that if and when a nucleotide base shifted into its rarer tautomeric form (the "imino" or "enol" form), a likely result would be base-pair mismatching. But evidence for these types of tautomeric shifts remains sparse.

Figure 1: Tautomeric shifts in nucleotide bases.
The purine and pyrimidine bases in DNA exist in two different tautomers, or chemical forms. (A) Nucleotide bases shift from their common “keto” form to their rarer, tautomeric “enol” form. (B) In common base pair arrangements, the common form of thymine (T) binds with the common form of adenine (A), and the common form of cytosine (C) binds with the common form of guanine (G). (C) Rare base-pairing arrangements result when one nucleotide in a base pair is the rare form instead of the common form. Here, the rare form of cytosine binds to the common form of adenine instead of guanine. The rare form of guanine binds to the common form of thymine instead of cytosine.
© 2014 Nature Education Adapted from Pierce, Benjamin. Genetics: A Conceptual Approach, 2nd ed. All rights reserved. 
Today, scientists suspect that most DNA replication errors are caused by mispairings of a different nature: either between different but nontautomeric chemical forms of bases (e.g., bases with an extra proton, which can still bind but often with a mismatched nucleotide, such as an A with a G instead of a T) or between "normal" bases that nonetheless bond inappropriately (e.g., again, an A with a G instead of a T) because of a slight shift in position of the nucleotides in space (Figure 2). This type of mispairing is known as wobble. It occurs because the DNA double helix is flexible and able to accommodate slightly misshaped pairings (Crick, 1966).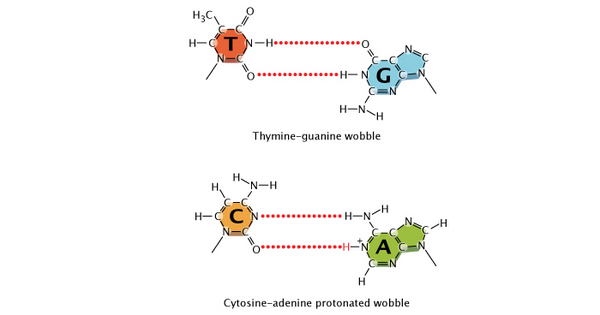

Figure 2: Wobble in mismatched nucleotide base pairs.
A shift in the position of nucleotides causes a wobble between a normal thymine and normal guanine. An additional proton on adenine causes a wobble in an adenine-cytosine base-pair.
© Nature Education Adapted from Pierce, Benjamin. Genetics: A Conceptual Approach, 2nd ed. All rights reserved. 
Replication errors can also involve insertions or deletions of nucleotide bases that occur during a process called strand slippage. Sometimes, a newly synthesized strand loops out a bit, resulting in the addition of an extra nucleotide base (Figure 3). Other times, the template strand loops out a bit, resulting in the omission, or deletion, of a nucleotide base in the newly synthesized, or primer, strand. Regions of DNA containing many copies of small repeated sequences are particularly prone to this type of error.