The postulated 99.9% genetic identicalness of all humans has been recently called into question due to an improved understanding of structural variations in human DNA. While most initial studies of genetic variation concentrated on individual nucleotide sequences, investigators have also found that large-scale changes occur in many locations throughout the genome. These insertions, deletions, inversions, and duplications result in changes in the physical arrangement of genes on chromosomes. Indeed, for as long as cytogeneticists have studied chromosomes under microscopes, they have observed variations in chromosomal structure. These scientists have noted such anomalies as aneuploidy (abnormal chromosome number), translocations of material from one chromosome to another, large-scale deletions and insertions, fragile sites, and variations in the size of the Y chromosome (Feuk et al., 2006).
The duplication of the Bar gene in Drosophila was one of the earliest structural variations to be linked to a phenotype. Seventy years ago, this variation was shown to cause the eye field of affected flies to be much narrower than that of flies with wild-type eyes (Bridges, 1936). In yet another example of a phenotypic link to a chromosomal anomaly, in humans, the duplication of part or all of chromosome 21 has been associated with Down syndrome. This duplication may be the result of nondisjunction or of translocation.
More recently, both aneuploidy and chromosomal translocations have been causally implicated in human cancers. These changes usually arise in individual somatic cells. While the study of such structural variations was initially limited to individual changes that could be seen through light microscopes, the advent of new technologies has allowed identification of submicroscopic structural variations on a genome-wide scale.
Discovering and Defining CNVs
© 2006 Nature Publishing Group Redon, R. et al. Global variation in copy number in the human genome. Nature 444, 447 (2006). All rights reserved. 
CNVs have been found in all human populations, as well as in other mammalian species (Freeman et al., 2006). Perhaps the best-defined and most widely known CNVs are the trinucleotide repeats (TNRs), which consist of three nucleotides repeating in tandem. TNRs exhibit dynamic expansion and contraction in a number of disease states, such as fragile X syndrome and Huntington's disease, with the number of repeats varying in both normal and afflicted individuals. In most cases, TNRs exhibit expansion with age. Because they are inherited through families, increased copy numbers typically correlate with greater disease severity and/or earlier onset of symptoms. Contraction of TNRs has been observed less frequently than expansion, typically upon paternal inheritance (Pearson et al., 2005).
Recently, a collaboration of international research laboratories has begun compiling a complete catalog of existing CNVs in the human genome. An examination of 270 DNA samples from the multiethnic population employed by the HapMap Project revealed a total of 1,447 discrete CNVs. Taken together, these CNVs cover approximately 360 megabases, or 12% of the human genome. The HapMap Project notes that CNVs encompass more nucleotide content per genome than SNPs, underscoring CNVs' significance to genetic diversity. A genome-wide map of CNVs shows that no region of the genome is exempt, and that the percentage of an individual's chromosomes that exhibit CNVs varies anywhere from 6% to 19% (Figure 1; Redon et al., 2006).

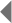




 Figure 1: Genomic distribution of copy number variable regions (CNVRs).
Figure 1: Genomic distribution of copy number variable regions (CNVRs).