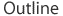How do researchers "read" gene sequences?
Determining the order of the nucleotides within a gene is known as DNA sequencing. The earliest DNA sequencing methods were time consuming, but a major breakthrough came in 1975 with the development of the process called Sanger sequencing. Sanger sequencing is named after English biochemist Frederick Sanger, and it is sometimes also referred to as chain-termination sequencing or dideoxy sequencing. Some 25 years after its creation, the Sanger method was used to sequence the human genome, and, with the addition of many technological improvements and modifications, it remains an important method in laboratories across the world today.
How does Sanger sequencing work?
Understanding DNA replication

Figure 1: DNA polymerase assembles nucleotides to make a new DNA strand.
Setting up the sequencing experiment
The Sanger method relies upon a variation of the replication process described above in order to determine the sequence of nucleotides in a segment of DNA. Before Sanger sequencing can begin, however, researchers must first make many copies of, or amplify, the DNA segment they wish to sequence. This is done either by cloning the DNA or by triggering the polymerase chain reaction (PCR). Once the DNA has been amplified, it is heated so that the two strands separate, and a synthetic primer is added to the mixture. The primer's sequence is complementary to the first piece of target DNA, which means that the primer and the DNA target bind with each other. At this point, the target sequence is exposed to a solution that contains DNA polymerase and all of the nucleotides required for synthesis of the complementary DNA strand — along with one special ingredient.
Adding ddNTPs
As described above, the next major step in the Sanger process is to expose the target sequence to DNA polymerase and significant amounts of all four nucleotides. In their unbound form, nucleotides have three phosphate groups and are formally called deoxynucleotide triphosphates, or dNTPs (where the "N" is a placeholder for A, T, G, or C). During the construction of a new DNA strand, a molecule called a hydroxyl group (which contains an oxygen atom and a hydrogen atom) attaches to the sugar of the last dNTP in the strand and chemically binds to the phosphate group on the next dNTP. This binding causes the DNA chain to grow. In Sanger sequencing, however, a special type of "dummy" nucleotide is included with the regular dNTPs that surround the growing DNA strand. These special nucleotides are known as dideoxynucleotide triphosphates, or ddNTPs (Figure 2), and they lack the crucial hydroxyl group that is attached to the sugar of dNTPs. Therefore, whenever a ddNTP is added to a growing DNA strand, it is unable to chemically bind with the next nucleotide in the chain, and the DNA strand stops growing.
When researchers carry out the Sanger process, they are manipulating many copies of the template strand at once, so an overabundance of dNTPs is required in order for DNA synthesis to proceed unimpeded on these copies until a ddNTP is added. Then, after the supply of dNTPs has been exhausted, the final result of the sequencing experiment is a group of new DNA strands of varying lengths. These strands all have a terminal ddNTP that indicates whether an A, T, G, or C occurs in that position on the template strand (Figure 3).
Reading the sequence: Now and then
When Sanger sequencing was first introduced, four separate reagents were used, one for each type of ddNTP. The four reaction products were then separated by gel electrophoresis, a process that organizes DNA fragments in order of size. This enabled researchers to assess the lengths of the truncated strands in each sample. This was important, because the end of each truncated strand was used to determine the position at which a ddNTP was added to the strand, thereby halting DNA elongation.
More recently, automation of the Sanger technique has made this process more efficient by combining all four ddNTP reactions in a single test tube. Each of the four ddNTPs in the tube is labeled with a different fluorescent color. Rather than being run on a gel and read manually, the reaction products are passed through a small tube containing a gel-like matrix. As the different-sized DNA fragments pass through the tube, a sequencing machine reads the fluorescent label at each position. Sequencing machines have vastly increased the speed and efficiency of DNA sequencing, and this technology continues to evolve at an astonishing rate.
How is DNA sequencing used by scientists?
In recent years, DNA sequencing technology has advanced many areas of science. For example, the field of functional genomics is concerned with figuring out what certain DNA sequences do, as well as which pieces of DNA code for proteins and which have important regulatory functions. An invaluable first step in making these determinations is learning the nucleotide sequences of the DNA segments under study. Another area of science that relies heavily on DNA sequencing is comparative genomics, in which researchers compare the genetic material of different organisms in order to learn about their evolutionary history and degree of relatedness. DNA sequencing has also aided complex disease research by allowing scientists to catalogue certain genetic variations between individuals that may influence their susceptibility to different conditions.
How can all people benefit from DNA sequencing?
More about sequencing