Abstract
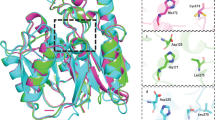
Nitric oxide synthase is inhibited by asymmetric NG-methylated derivatives of arginine whose cellular levels are controlled in part by dimethylarginine dimethylaminohydrolase (DDAH, EC 3.5.3.18). Levels of asymmetric NG,NG-dimethylarginine (ADMA) are known to correlate with certain disease states. Here, the first structure of a DDAH shows an unexpected similarity to arginine:glycine amidinotransferase (EC 2.1.4.1) and arginine deiminase (EC 3.5.3.6), thus defining a superfamily of arginine-modifying enzymes. The identification of a Cys-His-Glu catalytic triad and the structures of a Cys to Ser point mutant bound to both substrate and product suggest a reaction mechanism. Comparison of the ADMA–DDAH and arginine–amidinotransferase complexes reveals a dramatic rotation of the substrate that effectively maintains the orientation of the scissile bond of the substrate with respect to the catalytic residues. The DDAH structure will form a basis for the rational design of selective inhibitors, which are of potential use in modulating NO synthase activity in pathological settings.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Colasanti, M. & Suzuki, H. TIPS 21, 249–252 (2000).
Fischmann, T.O. et al. Nature Struct. Biol. 6, 233–242 (1999).
Moncada, S. J. Roy. Soc. Med. 92, 164–169 (1999).
Leiper, J.M. & Vallance, P. Cardiovasc. Res. 43, 542–548 (1999).
Vallance, P., Leone, A., Calver, A., Collier, J. & Moncada, S. Lancet 339, 572–575 (1992).
Miyazaki, H. et al. Circulation 99, 1141–1146 (1999).
MacAllister, R.J. et al. British J. Pharm. 119, 1533–1540 (1996).
Leiper, J.M. et al. Biochem. J. 343, 209–214 (1999).
Tran, C.T.L., Fox, M.F., Vallance, P. & Leiper, J.M. Genomics 67, 101–105 (2000).
Santa Maria, J., Vallance, P., Charles, I.G. & Leiper, J.M. Mol. Microbiol. 33, 1278–1279 (1999).
Humm, A., Fritsche, E., Steinbacher, S. & Huber, R. EMBO J. 16, 3373–3385 (1997).
Fritsche E., Humm A. & Huber R. J. Biol .Chem. 274, 3026–3032 (1999).
Fritsche, E., Bergner, A., Humm, A., Piepersberg, W. & Huber, R. Biochemistry 37, 17664–17672 (1998).
Ogawa, T., Kimoto, M. & Sasaoka, K. J. Biol. Chem. 264, 10205–10209 (1989).
Storer, A.C. & Ménard, R. Methods Enzymol. 244, 486–500 (1994).
Knodler, L.A., Sekyere, E.O., Stewart, T.S., Schofield, P.J. & Edwards, M.R. J. Biol. Chem. 273, 4470–4477 (1998).
Hendrickson, W.A., Horton, J.R. & LeMaster, D.M. EMBO J. 9, 1665–1672 (1990).
Leslie A.G.W. Joint CCP4 and ESF-EAMCB Newsletter on Protein Crystallography 26 (1992.
CCP4 Collaborative Computing Project, Number 4. Acta Crystallogr. D 50, 760–763 (1994).
Terwilliger, T.C. & Berendzen, J. Acta Crystallogr. D 55, 849–861 (1999).
Cowtan, K. In Joint CCP4 and ESF-EACBM Newsletter on Protein Crystallography 31, 34–38 (1994).
Navaza, J. Acta Crystallogr. A 50, 157–163 (1994).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Acta Crystallogr. A 47, 110–119 (1991).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Acta Crystallogr. D 53, 240–255 (1997).
Kraulis, P.J. J. Appl. Crystallogr. 24, 946–950 (1991).
Esnouf R.M. Acta Crystallogr. D 55, 938–940 (1999).
Acknowledgements
This work was initiated and funded in part by a British Heart Foundation Programme Grant to P.V., and by the BBSRC through the Bloomsbury Centre for Structural Biology. We thank ESRF and SRS synchrotron sources for use of data collection facilities, and P. Driscoll, D. Selwood and P. Murray-Rust for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Murray-Rust, J., Leiper, J., McAlister, M. et al. Structural insights into the hydrolysis of cellular nitric oxide synthase inhibitors by dimethylarginine dimethylaminohydrolase. Nat Struct Mol Biol 8, 679–683 (2001). https://doi.org/10.1038/90387
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/90387
This article is cited by
-
Determination of equilibria constants of arginine:glycine amidinotransferase (AGAT)-catalyzed reactions using concentrations of circulating amino acids
Amino Acids (2023)
-
Pharmacological activation of dimethylarginine dimethylaminohydrolase (DDAH) activity by inorganic nitrate and DDAH inhibition by NG-hydroxy-l-arginine, Nω,Nω-dimethyl-l-citrulline and Nω,Nω-dimethyl-Nδ-hydroxy-l-citrulline: results and overview
Amino Acids (2019)
-
Effects of single and combined metformin and l-citrulline supplementation on l-arginine-related pathways in Becker muscular dystrophy patients: possible biochemical and clinical implications
Amino Acids (2018)
-
Genetic variations in the alanine–glyoxylate aminotransferase 2 (AGXT2) gene and dimethylarginines levels in rheumatoid arthritis
Amino Acids (2017)
-
Retinal metabolic events in preconditioning light stress as revealed by wide-spectrum targeted metabolomics
Metabolomics (2017)