Abstract
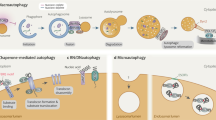
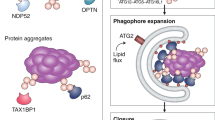
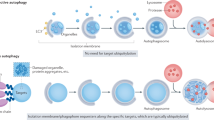
Autophagy complements the ubiquitin-proteasome system in mediating protein turnover. Whereas the proteasome degrades individual proteins modified with ubiquitin chains, autophagy degrades many proteins and organelles en masse. Macromolecules destined for autophagic degradation are 'selected' through sequestration within a specialized double-membrane compartment termed the phagophore, the precursor to an autophagosome, and then are hydrolyzed in a lysosome- or vacuole-dependent manner. Notably, a pair of distinctive ubiquitin-like proteins (UBLs), Atg8 and Atg12, regulate degradation by autophagy in unique ways by controlling autophagosome biogenesis and recruitment of specific cargos during selective autophagy. Here we review structural mechanisms underlying the functions and conjugation of these UBLs that are specialized to provide interaction platforms linked to phagophore membranes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Yang, Z. & Klionsky, D.J. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12, 814–822 (2010).
Reggiori, F. & Klionsky, D.J. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics 194, 341–361 (2013).
Mijaljica, D. et al. Receptor protein complexes are in control of autophagy. Autophagy 8, 1701–1705 (2012).
Oku, M. & Sakai, Y. Peroxisomes as dynamic organelles: autophagic degradation. FEBS J. 277, 3289–3294 (2010).
Kanki, T., Klionsky, D.J. & Okamoto, K. Mitochondria autophagy in yeast. Antioxid. Redox Signal. 14, 1989–2001 (2011).
Youle, R.J. & Narendra, D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14 (2011).
Knodler, L.A. & Celli, J. Eating the strangers within: host control of intracellular bacteria via xenophagy. Cell. Microbiol. 13, 1319–1327 (2011).
Geng, J. & Klionsky, D.J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. EMBO Rep. 9, 859–864 (2008).
Ichimura, Y. et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000).Identifies the Atg8–PE conjugation system.
Mizushima, N. et al. A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998).Identifies the Atg12–Atg5 conjugation system.
Weidberg, H. et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792–1802 (2010).
Xie, Z., Nair, U. & Klionsky, D.J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 19, 3290–3298 (2008).Shows that Atg8 levels correlate with the size of autophagosomes.
Nakatogawa, H., Ichimura, Y. & Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 (2007).
Nair, U. et al. SNARE proteins are required for macroautophagy. Cell 146, 290–302 (2011).
Nair, U. et al. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy 8, 780–793 (2012).
Shintani, T., Huang, W.-P., Stromhaug, P.E. & Klionsky, D.J. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev. Cell 3, 825–837 (2002).
Hanada, T. et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol. Chem. 282, 37298–37302 (2007).Identifies the Atg12–Atg5 complex as a new E3 ligase promoting Atg8 lipidation.
Kuma, A., Mizushima, N., Ishihara, N. & Ohsumi, Y. Formation of the approximately 350-kDa Apg12-Apg5•Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 277, 18619–18625 (2002).
Fujita, N. et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 19, 2092–2100 (2008).
Kraft, C. et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 31, 3691–3703 (2012).
Noda, N.N., Ohsumi, Y. & Inagaki, F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584, 1379–1385 (2010).
Noda, N.N. et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13, 1211–1218 (2008).Refs. 22 and 44 identify the structural basis for AIM or LIR binding to Atg8 (LC3) UBLs.
Behrends, C., Sowa, M.E., Gygi, S.P. & Harper, J.W. Network organization of the human autophagy system. Nature 466, 68–76 (2010).A proteomic analysis of Atg8 (LC3) family–interacting proteins, suggesting the complexity of the autophagy network.
Noda, N.N. et al. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol. Cell 44, 462–475 (2011).Refs. 24, 69 and 70 revealed mechanisms by which Atg7 initiates conjugation of autophagy UBLs.
Satoo, K. et al. The structure of Atg4B–LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 28, 1341–1350 (2009).Revealed the structural mechanisms underlying ATG4-dependent processing and deconjugation of LC3 family members.
Suzuki, H. et al. Structural basis of the autophagy-related LC3/Atg13 LIR complex: recognition and interaction mechanism. Structure 22, 47–58 (2014).
Pankiv, S. et al. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188, 253–269 (2010).
Seillier, M. et al. TP53INP1, a tumor suppressor, interacts with LC3 and ATG8-family proteins through the LC3-interacting region (LIR) and promotes autophagy-dependent cell death. Cell Death Differ. 19, 1525–1535 (2012).
Colecchia, D. et al. MAPK15/ERK8 stimulates autophagy by interacting with LC3 and GABARAP proteins. Autophagy 8, 1724–1740 (2012).
Popovic, D. et al. Rab GTPase-activating proteins in autophagy: regulation of endocytic and autophagy pathways by direct binding to human ATG8 modifiers. Mol. Cell. Biol. 32, 1733–1744 (2012).
Nowak, J. et al. The TP53INP2 protein is required for autophagy in mammalian cells. Mol. Biol. Cell 20, 870–881 (2009).
Birgisdottir, Å.B., Lamark, T. & Johansen, T. The LIR motif: crucial for selective autophagy. J. Cell Sci. 126, 3237–3247 (2013).
Lynch-Day, M.A. & Klionsky, D.J. The Cvt pathway as a model for selective autophagy. FEBS Lett. 584, 1359–1366 (2010).
Kanki, T., Wang, K., Cao, Y., Baba, M. & Klionsky, D.J. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17, 98–109 (2009).
Okamoto, K., Kondo-Okamoto, N. & Ohsumi, Y. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87–97 (2009).
Motley, A.M., Nuttall, J.M. & Hettema, E.H. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 31, 2852–2868 (2012).
Shaid, S., Brandts, C.H., Serve, H. & Dikic, I. Ubiquitination and selective autophagy. Cell Death Differ. 20, 21–30 (2013).
Pankiv, S. et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 (2007).
Kirkin, V. et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell 33, 505–516 (2009).
Thurston, T.L., Ryzhakov, G., Bloor, S., von Muhlinen, N. & Randow, F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 10, 1215–1221 (2009).
Johansen, T. & Lamark, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296 (2011).
Korac, J. et al. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J. Cell Sci. 126, 580–592 (2013).
Jiang, S., Wells, C.D. & Roach, P.J. Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1. Biochem. Biophys. Res. Commun. 413, 420–425 (2011).
Ichimura, Y. et al. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283, 22847–22857 (2008).
Rozenknop, A. et al. Characterization of the interaction of GABARAPL-1 with the LIR motif of NBR1. J. Mol. Biol. 410, 477–487 (2011).
Kaufmann, A., Beier, V., Franquelim, H.G. & Wollert, T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell 156, 469–481 (2014).
Birmingham, C.L., Smith, A.C., Bakowski, M.A., Yoshimori, T. & Brumell, J.H. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 281, 11374–11383 (2006).
Thurston, T.L., Wandel, M.P., von Muhlinen, N., Foeglein, A. & Randow, F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414–418 (2012).
von Muhlinen, N. et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol. Cell 48, 329–342 (2012).Revealed basis for LC3C specificity toward a CLIR.
Wild, P. et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 (2011).Refs. 50 and 51 demonstrated phosphorylation-dependent recruitment of cargo to Atg8 (LC3) family members.
Rogov, V.V. et al. Structural basis for phosphorylation-triggered autophagic clearance of Salmonella. Biochem. J. 454, 459–466 (2013).
Weiergräber, O.H. et al. Ligand binding mode of GABAA receptor-associated protein. J. Mol. Biol. 381, 1320–1331 (2008).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Kirisako, T. et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 151, 263–276 (2000).
Li, M. et al. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J. Biol. Chem. 286, 7327–7338 (2011).
Kabeya, Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000).
Hemelaar, J., Lelyveld, V.S., Kessler, B.M. & Ploegh, H.L. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1–LC3, GABARAP, and Apg8L. J. Biol. Chem. 278, 51841–51850 (2003).
Kabeya, Y. et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 117, 2805–2812 (2004).
Mariño, G. et al. Human autophagins, a family of cysteine proteinases potentially implicated in cell degradation by autophagy. J. Biol. Chem. 278, 3671–3678 (2003).
Tanida, I., Ueno, T. & Kominami, E. Human light chain 3/MAP1LC3B is cleaved at its carboxyl-terminal Met121 to expose Gly120 for lipidation and targeting to autophagosomal membranes. J. Biol. Chem. 279, 47704–47710 (2004).
Yu, Z.Q. et al. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy 8, 883–892 (2012).
Kumanomidou, T. et al. The crystal structure of human Atg4b, a processing and de-conjugating enzyme for autophagosome-forming modifiers. J. Mol. Biol. 355, 612–618 (2006).
Sugawara, K. et al. Structural basis for the specificity and catalysis of human Atg4B responsible for mammalian autophagy. J. Biol. Chem. 280, 40058–40065 (2005).
Sakoh-Nakatogawa, M. et al. Atg12–Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat. Struct. Mol. Biol. 20, 433–439 (2013).
Noda, N.N., Fujioka, Y., Hanada, T., Ohsumi, Y. & Inagaki, F. Structure of the Atg12–Atg5 conjugate reveals a platform for stimulating Atg8–PE conjugation. EMBO Rep. 14, 206–211 (2013).Refs. 65 and 79 revealed the structure of the Atg12–Atg5 conjugate.
Radoshevich, L. et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 142, 590–600 (2010).
Tanida, I. et al. Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell 10, 1367–1379 (1999).
Schulman, B.A. & Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 10, 319–331 (2009).
Taherbhoy, A.M. et al. Atg8 transfer from Atg7 to Atg3: a distinctive E1–E2 architecture and mechanism in the autophagy pathway. Mol. Cell 44, 451–461 (2011).
Hong, S.B. et al. Insights into noncanonical E1 enzyme activation from the structure of autophagic E1 Atg7 with Atg8. Nat. Struct. Mol. Biol. 18, 1323–1330 (2011).
Kaiser, S.E. et al. Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7–Atg3 and Atg7–Atg10 structures. Nat. Struct. Mol. Biol. 19, 1242–1249 (2012).Refs. 71 and 72 revealed mechanisms of Atg7 interactions with the autophagy E2s Atg3 and Atg10.
Yamaguchi, M. et al. Noncanonical recognition and UBL loading of distinct E2s by autophagy-essential Atg7. Nat. Struct. Mol. Biol. 19, 1250–1256 (2012).
Komatsu, M. et al. The C-terminal region of an Apg7p/Cvt2p is required for homodimerization and is essential for its E1 activity and E1–E2 complex formation. J. Biol. Chem. 276, 9846–9854 (2001).
Hong, S.B., Kim, B.W., Kim, J.H. & Song, H.K. Structure of the autophagic E2 enzyme Atg10. Acta Crystallogr. D Biol. Crystallogr. 68, 1409–1417 (2012).
Yamada, Y. et al. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J. Biol. Chem. 282, 8036–8043 (2007).
Yamaguchi, M. et al. Structural insights into Atg10-mediated formation of the autophagy-essential Atg12-Atg5 conjugate. Structure 20, 1244–1254 (2012).
Cao, Y., Cheong, H., Song, H. & Klionsky, D.J. In vivo reconstitution of autophagy in Saccharomyces cerevisiae. J. Cell Biol. 182, 703–713 (2008).
Romanov, J. et al. Mechanism and functions of membrane binding by the Atg5–Atg12/Atg16 complex during autophagosome formation. EMBO J. 31, 4304–4317 (2012).
Otomo, C., Metlagel, Z., Takaesu, G. & Otomo, T. Structure of the human ATG12∼ATG5 conjugate required for LC3 lipidation in autophagy. Nat. Struct. Mol. Biol. 20, 59–66 (2012).
Metlagel, Z., Otomo, C., Takaesu, G. & Otomo, T. Structural basis of ATG3 recognition by the autophagic ubiquitin-like protein ATG12. Proc. Natl. Acad. Sci. USA 110, 18844–18849 (2013).
Brownell, J.E. et al. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol. Cell 37, 102–111 (2010).
Boada-Romero, E. et al. TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3. EMBO J. 32, 566–582 (2013).
Travassos, L.H. et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat. Immunol. 11, 55–62 (2010).
Nakagawa, I. et al. Autophagy defends cells against invading group A Streptococcus. Science 306, 1037–1040 (2004).
Hwang, S. et al. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe 11, 397–409 (2012).
Cadwell, K., Patel, K.K., Komatsu, M., Virgin, H.W. IV & Stappenbeck, T.S. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy 5, 250–252 (2009).
Tanji, K., Mori, F., Kakita, A., Takahashi, H. & Wakabayashi, K. Alteration of autophagosomal proteins (LC3, GABARAP and GATE-16) in Lewy body disease. Neurobiol. Dis. 43, 690–697 (2011).
Chen, D. et al. Genetic analysis of the ATG7 gene promoter in sporadic Parkinson’s disease. Neurosci. Lett. 534, 193–198 (2013).
Chen, D. et al. A novel and functional variant within the ATG5 gene promoter in sporadic Parkinson’s disease. Neurosci. Lett. 538, 49–53 (2013).
Metzger, S. et al. Age at onset in Huntington’s disease is modified by the autophagy pathway: implication of the V471A polymorphism in Atg7. Hum. Genet. 128, 453–459 (2010).
Komatsu, M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005).
Ding, W.X. et al. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology 139, 1740–1752 (2010).
Takamura, A. et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 25, 795–800 (2011).
Hara, T. et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006).Refs. 94 and 95 show that neuron-specific loss of ATG5 or ATG7 leads to neurodegeneration.
Komatsu, M. et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 (2006).
Masiero, E. et al. Autophagy is required to maintain muscle mass. Cell Metab. 10, 507–515 (2009).
Raben, N. et al. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum. Mol. Genet. 17, 3897–3908 (2008).
Saitoh, T. et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456, 264–268 (2008).
Cadwell, K. et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 456, 259–263 (2008).
Hampe, J. et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 39, 207–211 (2007).
Rioux, J.D. et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat. Genet. 39, 596–604 (2007).
Novak, I. et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11, 45–51 (2010).Refs. 102 and 103 show that BNIP3L (Nix) is a mitophagy receptor that binds LC3.
Sandoval, H. et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature 454, 232–235 (2008).
Kuma, A. et al. The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004).
Tsukamoto, S. et al. Autophagy is essential for preimplantation development of mouse embryos. Science 321, 117–120 (2008).
Mortensen, M. et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J. Exp. Med. 208, 455–467 (2011).
Kitamura, K. et al. Autophagy-related Atg8 localizes to the apicoplast of the human malaria parasite Plasmodium falciparum. PLoS ONE 7, e42977 (2012).
Scherz-Shouval, R. et al. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749–1760 (2007).
Nair, U., Cao, Y., Xie, Z. & Klionsky, D.J. Roles of the lipid-binding motifs of Atg18 and Atg21 in the cytoplasm to vacuole targeting pathway and autophagy. J. Biol. Chem. 285, 11476–11488 (2010).
Thompson, A.R., Doelling, J.H., Suttangkakul, A. & Vierstra, R.D. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 138, 2097–2110 (2005).
Mizushima, N. et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657–668 (2001).
Hain, A.U.P. et al. Structural characterization and inhibition of the Plasmodium Atg8-Atg3 interaction. J. Struct. Biol. 180, 551–562 (2012).
Yamaguchi, M. et al. Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J. Biol. Chem. 285, 29599–29607 (2010).
Martinez, J. et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl. Acad. Sci. USA 108, 17396–17401 (2011).
Ishibashi, K., Uemura, T., Waguri, S. & Fukuda, M. Atg16L1, an essential factor for canonical autophagy, participates in hormone secretion from PC12 cells independently of autophagic activity. Mol. Biol. Cell 23, 3193–3202 (2012).
Yousefi, S. et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 8, 1124–1132 (2006).
Lee, I. & Schindelin, H. Structural insights into E1-catalyzed ubiquitin activation and transfer to conjugating enzymes. Cell 134, 268–278 (2008).
Acknowledgements
We thank S. Kaiser, M. Yamaguchi and Y. Qiu for helpful comments. The Klionsky laboratory is supported by US National Institutes of Health (NIH) grant 2R01GM053396. Research on autophagy in the Schulman laboratory is supported by the American Lebanese Syrian Associated Charities, the Howard Hughes Medical Institute, the St. Jude Cancer Center grant 5P30CA021765 and NIH grant R01GM077053.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Klionsky, D., Schulman, B. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Mol Biol 21, 336–345 (2014). https://doi.org/10.1038/nsmb.2787
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2787
This article is cited by
-
Finding New Targets for the Treatment of Heart Failure: Endoplasmic Reticulum Stress and Autophagy
Journal of Cardiovascular Translational Research (2023)
-
Golgi-associated Rab GTPases implicated in autophagy
Cell & Bioscience (2021)
-
Membrane perturbation by lipidated Atg8 underlies autophagosome biogenesis
Nature Structural & Molecular Biology (2021)
-
Genome-wide analysis of autophagy-related genes in Medicago truncatula highlights their roles in seed development and response to drought stress
Scientific Reports (2021)
-
The Effect and Mechanism of Asymmetric Dimethylarginine Regulating Trophoblastic Autophagy on Fetal Growth Restriction
Reproductive Sciences (2021)