Abstract
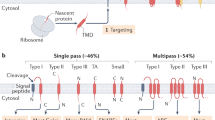
The membrane protein–folding problem can be articulated by two central questions. How is protein topology established by selective peptide transport to opposite sides of the cellular membrane? And how are transmembrane segments inserted, integrated and folded within the lipid bilayer? In eukaryotes, this process usually takes place in the endoplasmic reticulum, coincident with protein synthesis, and is facilitated by the translating ribosome and the Sec61 translocon complex (RTC). At its core, the RTC forms a dynamic pathway through which the elongating nascent polypeptide moves as it is delivered into the cytosolic, lumenal and lipid compartments. This Perspective will focus on emerging evidence that the RTC functions as a protein-folding machine that restricts conformational space by establishing transmembrane topology and yet provides a permissive environment that enables nascent transmembrane domains to efficiently progress down their folding energy landscape.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Popot, J.L. & Engelman, D. Helical membrane protein folding, stability and evolution. Annu. Rev. Biochem. 69, 881–922 (2000).
Bowie, J.U. Solving the membrane protein folding problem. Nature 438, 581–589 (2005).
Johnson, A.E. & van Waes, M. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 15, 799–842 (1999).
Rapoport, T.A., Goder, V., Heinrich, S. & Matlack, K. Membrane-protein integration and the role of the translocation channel. Trends Cell Biol. 14, 568–575 (2004).
Blobel, G. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA 77, 1496–1500 (1980).
Schnell, D.J. & Hebert, D. Protein translocons: multifunctional mediators of protein translocation across membranes. Cell 112, 491–505 (2003).
White, S.H. & von Heijne, G. The machinery of membrane protein assembly. Curr. Opin. Struct. Biol. 14, 397–404 (2004).
Johnson, A. Fluorescence approaches for determining protein conformations, interactions and mechanisms at membranes. Traffic 6, 1078–1092 (2005).
von Heijne, G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 7, 909–918 (2006).
Osborne, A.R., Rapoport, T.A. & van den Berg, B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell Dev. Biol. 21, 529–550 (2005).
Pitonzo, D. & Skach, W. Molecular mechanisms of aquaporin biogenesis by the endoplasmic reticulum Sec61 translocon. Biochim. Biophys. Acta 1758, 976–988 (2006).
Sadlish, H. & Skach, W. Biogenesis of CFTR and other polytopic membrane proteins; new roles for the ribosome-translocon complex. J. Membr. Biol. 202, 115–126 (2004).
Walter, P. & Lingappa, V.R. Mechanisms of protein translocation across the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 2, 499–516 (1986).
Song, W., Raden, D., Mandon, E. & Gilmore, R. Role of Sec61α in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell 100, 333–343 (2000).
Görlich, D., Hartmann, E., Prehn, S. & Rapoport, T. A protein of the endoplasmic reticulum involved in early polypeptide translocation. Nature 357, 47–52 (1992).
Wiedmann, M., Kurzchalia, T., Hartmann, E. & Rapoport, T. A signal sequence receptor in the endoplasmic reticulum membrane. Nature 328, 830–833 (1987).
Kelleher, D.J. & Gilmore, R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47R–62R (2006).
Shibatani, T., David, L., McCormack, A., Frueh, K. & Skach, W. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP and two potential new subunits. Biochemistry 44, 5982–5992 (2005).
Snapp, E.L., Reinhart, G., Bogert, B., Lippincott-Schwartz, J. & Hegde, R. The organization of engaged and quiescent translocons in the endoplasmic reticulum of mammalian cells. J. Cell Biol. 164, 997–1007 (2004).
van den Berg, B. et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004).
Ménétret, J.-F. et al. Architecture of the ribosome-channel complex derived from native membranes. J. Mol. Biol. 348, 445–457 (2005).
Ménétret, J.F. et al. Single copies of Sec61 and TRAP associate with a nontranslating mammalian ribosome. Structure 16, 1126–1137 (2008).
Beckmann, R. et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107, 361–372 (2001).
Belin, D., Bost, S., Vassalli, J.-D. & Strub, K. A two step recognition of signal sequences determines the translocation efficiency of proteins. EMBO J. 15, 468–478 (1996).
Jungnickel, B. & Rapoport, T. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell 82, 261–270 (1995).
Crowley, K.S., Liao, S., Worrell, V., Reinhart, G. & Johnson, A. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell 78, 461–471 (1994).
Crowley, K.S., Reinhart, G. & Johnson, A. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell 73, 1101–1115 (1993).
Kim, S.J., Mitra, D., Salerno, J. & Hegde, R. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev. Cell 2, 207–217 (2002).
Hegde, R.S. & Kang, S.-W. The concept of translocational regulation. J. Cell Biol. 182, 225–232 (2008).
Pitonzo, D., Yang, Z., Matsumura, Y., Johnson, A. & Skach, W. Sequence-specific retention and regulated integration of a nascent membrane protein by the ER Sec61 translocon. Mol. Biol. Cell 20, 685–698 (2009).
Woolhead, C.A., McCormick, P. & Johnson, A. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116, 725–736 (2004).
Alder, N.N., Shen, Y., Brodsky, J., Hendershot, L. & Johnson, A. The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J. Cell Biol. 168, 389–399 (2005).
Haigh, N.G. & Johnson, A. A new role for BiP: closing the aqueous translocon pore during protein integration into the ER membrane. J. Cell Biol. 156, 261–270 (2002).
Liao, S., Lin, J., Do, H. & Johnson, A. Both lumenal and cytosolic gating of the aqueous translocon pore are regulated from inside the ribosome during membrane protein integration. Cell 90, 31–41 (1997).
Martoglio, B., Hofmann, M., Brunner, J. & Dobberstein, B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell 81, 207–214 (1995).
Heinrich, S.U., Mothes, W., Brunner, J. & Rapoport, T. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell 102, 233–244 (2000).
Hessa, T. et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377–381 (2005).
Hessa, T. et al. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450, 1026–1030 (2007).
Enquist, K. et al. Membrane-integration characteristics of two ABC transporters, CFTR and P-glycoprotein. J. Mol. Biol. 387, 1153–1164 (2009).
Do, H., Falcone, D., Lin, J., Andrews, D. & Johnson, A. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell 85, 369–378 (1996).
Sadlish, H., Pitonzo, D., Johnson, A. & Skach, W. Sequential triage of transmembrane segments by Sec61α during biogenesis of a native multispanning membrane protein. Nat. Struct. Mol. Biol. 12, 870–878 (2005).
McCormick, P.J., Miao, Y., Shao, Y., Lin, J. & Johnson, A. Cotranslational protein integration into the ER membrane is mediated by the binding of nascent chains to translocon proteins. Mol. Cell 12, 329–341 (2003).
Meacock, S.L., Lecomte, F., Crawshaw, S. & High, S. Different transmembrane domains associate with distinct endoplasmic reticulum components during membrane integration of a polytopic protein. Mol. Biol. Cell 13, 4114–4129 (2002).
Ismail, N., Crawshaw, S., Cross, B., Haagsma, A. & High, S. Specific transmembrane segments are selectively delayed at the ER translocon during opsin biogenesis. Biochem. J. 411, 495–506 (2008).
Hamman, B.D., Hendershot, L. & Johnson, A. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell 92, 747–758 (1998).
Skach, W. Topology of P-glycoproteins. Methods Enzymol. 292, 265–278 (1998).
Xie, Y., Langhans-Rajasekaran, S., Bellovino, D. & Morimoto, T. Only the first and the last hydrophobic segments in the COOH-terminal third of the Na,K-ATPase α subunit initiate and halt, respectively, membrane translocation of the newly synthesized polypeptide. J. Biol. Chem. 271, 2563–2573 (1996).
Skach, W.R., Calayag, M.C. & Lingappa, V. Evidence for an alternate model of human P-glycoprotein structure and biogenesis. J. Biol. Chem. 268, 6903–6908 (1993).
Skach, W.R. & Lingappa, V. Amino terminus assembly of human P-glycoprotein at the endoplasmic reticulum is directed by cooperative actions of two internal sequences. J. Biol. Chem. 268, 23552–23561 (1993).
Skach, W.R. et al. Biogenesis and transmembrane topology of the CHIP28 water channel in the endoplasmic reticulum. J. Cell Biol. 125, 803–815 (1994).
Moss, K., Helm, A., Lu, Y., Bragin, A. & Skach, W. Coupled translocation events generate topologic heterogeneity at the endoplasmic reticulum membrane. Mol. Biol. Cell 9, 2681–2697 (1998).
Audigier, Y., Friedlander, M. & Blobel, G. Multiple topogenic sequences in bovine opsin. Proc. Natl. Acad. Sci. USA 84, 5783–5787 (1987).
Agre, P. et al. Aquaporin water channels—from atomic structure to clinical medicine. J. Physiol. (Lond.) 542, 3–16 (2002).
King, L.S., Kozono, D. & Agre, P. From structure to disease, the evolving tale of aquaporin biology. Nat. Rev. Mol. Cell Biol. 5, 687–698 (2004).
Fujiyoshi, Y. et al. Structure and function of water channels. Curr. Opin. Struct. Biol. 12, 509–515 (2002).
Sui, H., Han, B.-G., Lee, J., Walian, P. & Jap, B. Structural basis of water specific transport through the AQP1 water channel. Nature 414, 872–878 (2001).
Fu, D. et al. Structure of a glycerol-conducting channel and the basis for its selectivity. Science 290, 481–486 (2000).
Verbavatz, J.M. et al. Tetrameric assembly of CHIP28 water channels in liposomes and cell membranes. A freeze-fracture study. J. Cell Biol. 123, 605–618 (1993).
Smith, B.L. & Agre, P. Erythrocyte Mr 28,000 transmembrane protein exists as a multisubunit oligomer similar to channel proteins. J. Biol. Chem. 266, 6407–6415 (1991).
Buck, T.M., Wagner, J., Grund, S. & Skach, W. A novel tripartite structural motif involved in aquaporin topogenesis, monomer folding and tetramerization. Nat. Struct. Mol. Biol. 14, 762–769 (2007).
Shi, L.-B., Skach, W., Ma, T. & Verkman, A. Distinct biogenesis mechanisms for water channels MIWC and CHIP28 at the endoplasmic reticulum. Biochemistry 34, 8250–8256 (1995).
Lu, Y. et al. Reorientation of Aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol. Biol. Cell 11, 2973–2985 (2000).
Foster, W. et al. Identification of sequence determinants that direct different intracellular folding pathways for AQP1 and AQP4. J. Biol. Chem. 275, 34157–34165 (2000).
Tu, L.W., Wang, J., Helm, A., Skach, W. & Deutsch, C. Transmembrane biogenesis of Kv1.3. Biochemistry 39, 824–836 (2000).
Lu, Y. et al. Co- and posttranslational mechanisms direct CFTR N-terminus transmembrane assembly. J. Biol. Chem. 273, 568–576 (1998).
Gratkowski, H., Lear, J. & DeGrado, W. Polar side chains drive the association of model transmembrane peptides. Proc. Natl. Acad. Sci. USA 98, 880–885 (2001).
Zhou, F.X., Cocco, M., Russ, W., Brunger, A. & Engelman, D. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat. Struct. Biol. 7, 154–160 (2000).
Gafvelin, G. & von Heijne, G. Topological “frustration” in multispanning E. coli inner membrane. Cell 77, 401–412 (1994).
Goder, V., Bieri, C. & Spiess, M. Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J. Cell Biol. 147, 257–266 (1999).
Garrison, J., Kunkel, E., Hegde, R. & Taunton, J. A substrate-specific inhibitor of protein translocation into the endoplasmic reticulum. Nature 436, 285–289 (2005).
Bogdanov, M., Xie, J., Heacock, P. & Dowhan, W. To flip or not to flip: lipid-protein charge interactions are a determinant of final membrane protein topology. J. Cell Biol. 182, 925–935 (2008).
Brambillasca, S., Yabal, M., Makarow, M. & Borgese, N. Unassisted translocation of large polypeptide domains across phospholipd bilayers. J. Cell Biol. 175, 767–777 (2006).
Lin, J. & Addison, R. A novel integration signal that is composed of two transmembrane segments is required to integrate the Neurospora plasma membrane H+-ATPase into microsomes. J. Biol. Chem. 270, 6935–6941 (1995).
Ismail, N., Crawshaw, S. & High, S. Active and passive displacement of transmembrane domains during opsin biogenesis at the Sec61 translocon. J. Cell Sci. 119, 2826–2836 (2006).
Beckmann, R. et al. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science 278, 2123–2126 (1997).
Ménétret, J.F. et al. The structure of ribosome-channel complexes engaged in protein translocation. Mol. Cell 6, 1219–1232 (2000).
Hanein, D. et al. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell 87, 721–732 (1996).
Morgan, D.G., Menetret, J., Neuhof, A., Rapoport, T. & Akey, C. Structure of the mammalian ribosome-channel complex at 17Å resolution. J. Mol. Biol. 324, 871–886 (2002).
Young, J.C., Agashe, V., Siegers, K. & Hartl, F. Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5, 781–791 (2004).
Oberdorf, J., Pitonzo, D. & Skach, W. An energy-dependent maturation step is required for release of the cystic fibrosis transmembrane conductance regulator from early endoplasmic reticulum biosynthetic machinery. J. Biol. Chem. 280, 38193–38202 (2005).
Clemons, W.M., Menetret, J.-F., Akey, C. & Rapoport, T. Structural insight into the protein translocation channel. Curr. Opin. Struct. Biol. 14, 390–396 (2004).
Breyton, C., Haase, W., Rapoport, T., Kuehlbrandt, W. & Collinson, I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature 418, 662–665 (2002).
Mitra, K. et al. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438, 318–324 (2005).
Hamman, B.D., Chen, J.-C., Johnson, E. & Johnson, A. The aqueous pore through the translocon has a diameter of 40–60Å during cotranslational protein translocation at the ER membrane. Cell 89, 535–544 (1997).
Kida, Y., Morimoto, F. & Sakaguchi, M. Two translocating hydrophilic segments of a nascent chain span the ER membrane during multispanning protein topogenesis. J. Cell Biol. 179, 1441–1452 (2007).
Acknowledgements
I would like to thank B. Conti and other members of the Skach laboratory for helpful comments on this manuscript and the US National Institutes of Health (R01GM53457 and R01 DK51818) for supporting our research in this area.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Skach, W. Cellular mechanisms of membrane protein folding. Nat Struct Mol Biol 16, 606–612 (2009). https://doi.org/10.1038/nsmb.1600
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1600
This article is cited by
-
ABC-transporter CFTR folds with high fidelity through a modular, stepwise pathway
Cellular and Molecular Life Sciences (2023)
-
Aberrant expression of Sec61α in esophageal cancers
Journal of Cancer Research and Clinical Oncology (2019)
-
Codon bias and the folding dynamics of the cystic fibrosis transmembrane conductance regulator
Cellular & Molecular Biology Letters (2016)
-
A central cavity within the holo-translocon suggests a mechanism for membrane protein insertion
Scientific Reports (2016)
-
The calcium sensing receptor: from calcium sensing to signaling
Science China Life Sciences (2015)