Abstract
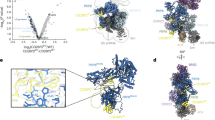
The U4/U6-U5 tri–small nuclear ribonucleoprotein (snRNP) is a major, evolutionarily highly conserved spliceosome subunit. Unwinding of its U4/U6 snRNA duplex is a central event of spliceosome activation that requires several components of the U5 portion of the tri-snRNP, including the RNA helicase Brr2, Prp8 and the GTPase Snu114. Here we report the EM projection structure of the Saccharomyces cerevisiae tri-snRNP. It shows a modular organization comprising three extruding domains that contact one another in its central portion. We have visualized genetically tagged tri-snRNP proteins by EM and show here that U4/U6 snRNP forms a domain termed the arm. Conversely, a separate head domain adjacent to the arm harbors Brr2, whereas Prp8 and the GTPase Snu114 are located centrally. The head and arm adopt variable relative positions. This molecular organization and dynamics suggest possible scenarios for structural events during catalytic activation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Will, C.L. & Lührmann, R. Spliceosome structure and function. in The RNA World (eds. Gesteland, R.F., Cech, T.R. & Atkins, J.F.) 369–400 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2006).
Deckert, J. et al. Protein composition and electron microscopy structure of affinity-purified human spliceosomal B complexes isolated under physiological conditions. Mol. Cell. Biol. 26, 5528–5543 (2006).
Gottschalk, A. et al. Identification by mass spectrometry and functional analysis of novel proteins of the yeast [U4/U6.U5] tri-snRNP. EMBO J. 18, 4535–4548 (1999).
Stevens, S.W. et al. Biochemical and genetic analyses of the U5, U6, and U4/U6 x U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA 7, 1543–1553 (2001).
Achsel, T., Ahrens, K., Brahms, H., Teigelkamp, S. & Lührmann, R. The human U5–220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 18, 6756–6766 (1998).
Kuhn, A.N., Reichl, E.M. & Brow, D.A. Distinct domains of splicing factor Prp8 mediate different aspects of spliceosome activation. Proc. Natl. Acad. Sci. USA 99, 9145–9149 (2002).
Small, E.C., Leggett, S.R., Winans, A.A. & Staley, J.P. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol. Cell 23, 389–399 (2006).
Galisson, F. & Legrain, P. The biochemical defects of prp4–1 and prp6–1 yeast splicing mutants reveal that the PRP6 protein is required for the accumulation of the [U4/U6.U5] tri-snRNP. Nucleic Acids Res. 21, 1555–1562 (1993).
Liu, S., Rauhut, R., Vornlocher, H.P. & Lührmann, R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA 12, 1418–1430 (2006).
Schaffert, N., Hossbach, M., Heintzmann, R., Achsel, T. & Lührmann, R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 23, 3000–3009 (2004).
Banroques, J. & Abelson, J.N. PRP4: a protein of the yeast U4/U6 small nuclear ribonucleoprotein particle. Mol. Cell. Biol. 9, 3710–3719 (1989).
Anthony, J.G., Weidenhammer, E.M. & Woolford, J.L., Jr. The yeast Prp3 protein is a U4/U6 snRNP protein necessary for integrity of the U4/U6 snRNP and the U4/U6.U5 tri-snRNP. RNA 3, 1143–1152 (1997).
Nottrott, S., Urlaub, H. & Lührmann, R. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. EMBO J. 21, 5527–5538 (2002).
Vidal, V.P., Verdone, L., Mayes, A.E. & Beggs, J.D. Characterization of U6 snRNA-protein interactions. RNA 5, 1470–1481 (1999).
Achsel, T. et al. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′- end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18, 5789–5802 (1999).
Nilsen, T.W. RNA-RNA interactions in nuclear pre-mRNA splicing. in RNA Structure and Function (ed. M.Grundber-Manago, R.W.S.A.) 279–307 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1998).
Staley, J.P. & Guthrie, C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92, 315–326 (1998).
Laggerbauer, B., Achsel, T. & Lührmann, R. The human U5–200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc. Natl. Acad. Sci. USA 95, 4188–4192 (1998).
Raghunathan, P.L. & Guthrie, C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr. Biol. 8, 847–855 (1998).
Kim, D.H. & Rossi, J.J. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. RNA 5, 959–971 (1999).
Grainger, R.J. & Beggs, J.D. Prp8 protein: at the heart of the spliceosome. RNA 11, 533–557 (2005).
Fabrizio, P., Laggerbauer, B., Lauber, J., Lane, W.S. & Lührmann, R. An evolutionarily conserved U5 snRNP-specific protein is a GTP-binding factor closely related to the ribosomal translocase EF-2. EMBO J. 16, 4092–4106 (1997).
Bartels, C., Klatt, C., Lührmann, R. & Fabrizio, P. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 3, 875–880 (2002).
Brenner, T.J. & Guthrie, C. Genetic analysis reveals a role for the C terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics 170, 1063–1080 (2005).
Bartels, C., Urlaub, H., Lührmann, R. & Fabrizio, P. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J. Biol. Chem. 278, 28324–28334 (2003).
Konarska, M.M., Vilardell, J. & Query, C.C. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol. Cell 21, 543–553 (2006).
Stark, H., Rodnina, M.V., Wieden, H.J., van Heel, M. & Wintermeyer, W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell 100, 301–309 (2000).
Puig, O. et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 (2001).
Kastner, B. et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 (2008).
Dube, P., Tavares, P., Lurz, R. & van Heel, M. The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J. 12, 1303–1309 (1993).
Sheff, M.A. & Thorn, K.S. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21, 661–670 (2004).
Campbell, R.E. et al. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99, 7877–7882 (2002).
Sander, B. et al. Organization of core spliceosomal components U5 snRNA loop I and U4/U6 Di-snRNP within U4/U6.U5 Tri-snRNP as revealed by electron cryomicroscopy. Mol. Cell 24, 267–278 (2006).
Lescoute, A. & Westhof, E. Topology of three-way junctions in folded RNAs. RNA 12, 83–93 (2006).
Liu, S. et al. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science 316, 115–120 (2007).
Jorgensen, R. et al. Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature 436, 979–984 (2005).
Jorgensen, R. et al. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat. Struct. Mol. Biol. 10, 379–385 (2003).
Chen, J.Y. et al. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD box splicing factor. Mol. Cell 7, 227–232 (2001).
Pyle, A.M. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 37, 317–336 (2008).
Gottschalk, A., Kastner, B., Lührmann, R. & Fabrizio, P. The yeast U5 snRNP coisolated with the U1 snRNP has an unexpected protein composition and includes the splicing factor Aar2p. RNA 7, 1554–1565 (2001).
Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 (1996).
Golas, M.M., Sander, B., Will, C.L., Lührmann, R. & Stark, H. Molecular architecture of the multiprotein splicing factor SF3b. Science 300, 980–984 (2003).
Sander, B., Golas, M.M. & Stark, H. Advantages of CCD detectors for de novo three-dimensional structure determination in single-particle electron microscopy. J. Struct. Biol. 151, 92–105 (2005).
Sander, B., Golas, M.M. & Stark, H. Corrim-based alignment for improved speed in single-particle image processing. J. Struct. Biol. 143, 219–228 (2003).
van Heel, M., Harauz, G., Orlova, E.V., Schmidt, R. & Schatz, M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 116, 17–24 (1996).
Acknowledgements
We thank H. Urlaub for MS analysis and M. Raabe for excellent technical assistance. We are grateful to C.L. Will for helpful comments on the manuscript and to R. Karaduman for tagging Prp8 and Snu114 with the TAP tag. This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie and the Ernst Jung Stiftung to R.L. Research in the laboratory of H.S. was supported by grants from the Federal Ministry of Education and Research, Germany (0311899) and the Sixth Framework Program of the European Union via the Integrated Project '3D Repertoire'. I.H. was supported by the Stiftung Stipendien-Fonds des Verbandes der Chemischen Industrie.
Author information
Authors and Affiliations
Contributions
I.H., B.S., M.M.G., P.F. and R.L. designed the experiments. P.F., H.S. and R.L. supervised the project. I.H. performed most of the biochemical and genetic research. B.S. and M.M.G. performed most of the EM research. E.W. performed the PA-gold labeling of the immunocomplexes and the EM of the PA-gold–labeled complexes. I.H. and E.W. analyzed the data of the PA-gold–labeled complexes. E.K. contributed to the biochemical and genetic experiments in the initial phase of the project. B.K. contributed to the EM research in the initial phase of the project. I.H., B.S., M.M.G., P.F., H.S. and R.L. analyzed the data and wrote the paper.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Methods (PDF 4223 kb)
Supplementary Video 1
Here, the two views show a repositioning of the head, which kinks to the left, accompanied by a small additional bending of the foot to the left, while the arm remains in the closed position. (GIF 54 kb)
Supplementary Video 2
In addition to the kinking of the head that is also evident in Supplementary Video 1, the arm changes between the closed and open position. No changes are observed in the foot region, while a slight bending of the central portion accompanies the changes in head and arm. (GIF 49 kb)
Rights and permissions
About this article
Cite this article
Häcker, I., Sander, B., Golas, M. et al. Localization of Prp8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy. Nat Struct Mol Biol 15, 1206–1212 (2008). https://doi.org/10.1038/nsmb.1506
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1506
This article is cited by
-
A de novo synonymous variant in EFTUD2 disrupts normal splicing and causes mandibulofacial dysostosis with microcephaly: case report
BMC Medical Genetics (2020)
-
The architecture of the spliceosomal U4/U6.U5 tri-snRNP
Nature (2015)
-
Strep-tag II and Twin-Strep Based Cassettes for Protein Tagging by Homologous Recombination and Characterization of Endogenous Macromolecular Assemblies in Saccharomyces cerevisiae
Molecular Biotechnology (2014)
-
Oto-facial syndrome and esophageal atresia, intellectual disability and zygomatic anomalies - expanding the phenotypes associated with EFTUD2 mutations
Orphanet Journal of Rare Diseases (2013)
-
Substrate binding on the APC/C occurs between the coactivator Cdh1 and the processivity factor Doc1
Nature Structural & Molecular Biology (2011)