Key Points
-
Matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI) mass spectrometry has become an important tool in the life sciences. Recently, mass spectrometry methods have been developed that are tailored to the rapid classification and identification of bacteria and other microorganisms.
-
MALDI-based protein mass pattern detection ('profiling') is an inexpensive and straightforward approach for bacterial classification and identification. The method can easily distinguish bacteria on the genus, species and, sometimes, subspecies level.
-
MALDI-based resequencing is an accurate, flexible and efficient alternative to conventional DNA sequencing applications in microbiology.
-
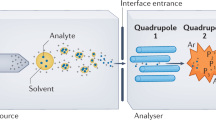
ESI-based detection of PCR products is a fast epidemiological tool for the analysis of microorganisms.
-
The three mass spectrometry approaches can be cleverly combined to provide comprehensive tools for bacterial detection.
-
Automated, standardized protocols and mature software packages for the mass spectrometry analysis of bacteria are available.
-
The methods can be easily adapted by microbiology laboratories using either academic or commercial protocols.
Abstract
Mass spectrometry has become an important analytical tool in biology in the past two decades. In principle, mass spectrometry offers high-throughput, sensitive and specific analysis for many applications in microbiology, including clinical diagnostics and environmental research. Recently, several mass spectrometry methods for the classification and identification of bacteria and other microorganisms, as well as new software analysis tools, have been developed. In this Review we discuss the application range of these mass spectrometry procedures and their potential for successful transfer into microbiology laboratories.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Peeling, R. W., Smith, P. G. & Bossuyt, P. M. A guide for diagnostic evaluations. Nature Rev. Microbiol. 4, S2–S6 (2006).
Coffey, A. G., Daly, C. & Fitzgerald, G. The impact of biotechnology on the dairy industry. Biotechnol. Adv. 12, 625–33 (1994).
Wilkinson, J. M. Silage and animal health. Nat. Toxins 7, 221–32 (1999).
Sintchenko, V., Iredell, J. R. & Gilbert, G. L. Pathogen profiling for disease management and surveillance. Nature Rev. Microbiol. 5, 464–470 (2007).
Holmes, B., Willcox, W. R. & Lapage, S. P. Identification of Enterobacteriaceae by the API 20E system. J. Clin. Pathol. 31, 22–30 (1978).
Engvall, E. Quantitative enzyme immunoassay (ELISA) in microbiology. Med. Biol. 55, 193–200 (1977).
O'Sullivan, T. F. & Fitzgerald, G. F. Comparison of Streptococcus thermophilus strains by pulse field gel electrophoresis of genomic DNA. FEMS Microbiol. Lett. 168, 213–219 (1998).
Sanger, F., Nickens, S. & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA 74, 5463–5467 (1977).
Shendure, J., Mitra, R. D., Varma, C. & Church, G. M. Advanced sequencing technologies: methods and goals. Nature Rev. Genet. 5, 335–344 (2004).
MacLean, D., Jones, J. D. & Studholme, D. J. Application of 'next-generation' sequencing technologies to microbial genetics. Nature Rev. Microbiol. 7, 287–296 (2009).
Gross, J. H. Mass Spectrometry – A Textbook (Springer, Heidelberg, 2004).
Hesse, M., Meier, H. & Zeeh, B. Spectroscopic Methods in Organic Chemistry 2nd edn (Thieme, Stuttgart, 2008).
Fox, A. Mass spectrometry for species or strain identification after culture or without culture: past, present, and future. J. Clin. Microbiol. 44, 2677–2680 (2006).
Karas, M. & Hillenkamp, F. Laser desorption ionization of proteins with molecular masses exceeding 10000 daltons. Anal. Chem. 60, 2299–2303 (1988).
Fenn, J. B. Electrospray wings for molecular elephants (Nobel lecture). Angew. Chem. Int. Edn Engl. 42, 3871–3894 (2003).
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Sauer, S. et al. Miniaturization in functional genomics and proteomics. Nature Rev. Genet. 6, 465–476 (2005).
Demirev, P. A., Ho, Y. P., Ryzhov, V. & Fenselau, C. Microorganism identification by mass spectrometry and protein database searches. Anal. Chem. 71, 2732–2738 (1999).
Freiwald, A. & Sauer, S. Phylogenetic classification and identification of bacteria by mass spectrometry. Nature Protoc. 4, 732–742 (2009). A practical guide for protein mass pattern analysis of bacteria.
Sauer, S. Typing of single nucleotide polymorphisms by MALDI mass spectrometry: principles and diagnostic applications. Clin. Chim. Acta 363, 95–105 (2006).
Ryzhov, V. & Fenselau, C. Characterization of the protein subset desorbed by MALDI from whole bacterial cells. Anal. Chem. 73, 746–750 (2001).
Teramoto, K., Sato, H., Sun, L., Torimura, M. & Tao, H. A simple intact protein analysis by MALDI-MS for characterization of ribosomal proteins of two genome-sequenced lactic acid bacteria and verification of their amino acid sequences. J. Proteome Res. 6, 3899–3907 (2007).
Claydon, M. A., Davey, S. N., Edwards-Jones, V. & Gordon, D. B. The rapid identification of intact microorganisms using mass spectrometry. Nature Biotech. 14, 1584–1586 (1996).
Holland, R. D. et al. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 10, 1227–1232 (1996). References 23 and 24 are the first papers to describe protein mass pattern detection of bacteria.
Chong, B. E., Wall, D. B., Lubman, D. M. & Flynn, S. J. Rapid profiling of E. coli proteins up to 500 kDa from whole cell lysates using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 11, 1900–1908 (1997).
Chen, P., Lu, Y. & Harrington, P. B. Biomarker profiling and reproducibility study of MALDI-MS measurements of Escherichia coli by analysis of variance-principal component analysis. Anal. Chem. 80, 1474–1481 (2008).
Williams, T. L., Andrzejewski, D., Lay, J. O. & Musser, S. M. Experimental factors affecting the quality and reproducibility of MALDI TOF mass spectra obtained from whole bacteria cells. J. Am. Soc. Mass Spectrom. 14, 342–351 (2003).
Fenselau, C. & Demirev, P. A. Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 20, 157–171 (2001).
Lay, J. O. Jr. MALDI-TOF mass spectrometry of bacteria. Mass Spectrom. Rev. 20, 172–194 (2001).
Lasch, P. et al. MALDI-TOF mass spectrometry compatible inactivation method for highly pathogenic microbial cells and spores. Anal. Chem. 80, 2026–2034 (2008).
Sauer, S. et al. Classification and identification of bacteria by mass spectrometry and computational analysis. PLoS ONE 3, e2843 (2008).
Bright, J. J., Claydon, M. A., Soufian, M. & Gordon, D. B. Rapid typing of bacteria using matrix-assisted laser desorption ionization time-of-flight mass spectrometry and pattern recognition software. J. Microbiol. Methods 48, 127–138 (2002).
Schmidt, F., Fiege, T., Hustoft, H. K., Kneist, S. & Thiede, B. Shotgun mass mapping of Lactobacillus species and subspecies from caries related isolates by MALDI-MS. Proteomics 9, 1994–2003 (2009).
Hsieh, S. Y. et al. Highly efficient classification and identification of human pathogenic bacteria by MALDI-TOF MS. Mol. Cell. Proteomics 7, 448–456 (2008).
Ilina, E. N. et al. Direct bacterial profiling by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry for identification of pathogenic Neisseria. J. Mol. Diagn. 11, 75–86 (2009).
Dieckmann, R., Helmuth, R., Erhard, M. & Malorny B. Rapid classification and identification of salmonellae at the species and subspecies levels by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74, 7767–7778 (2008).
Barbuddhe, S. B. et al. Rapid identification and typing of Listeria species by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 74, 5402–5407 (2008).
Grosse-Herrenthey, A. et al. Challenging the problem of clostridial identification with matrix-assisted laser desorption and ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Anaerobe 14, 242–249 (2008).
Carbonnelle, E. et al. Rapid identification of Staphylococci isolated in clinical microbiology laboratories by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 45, 2156–2161 (2007).
Teramoto, K. et al. Phylogenetic classification of Pseudomonas putida strains by MALDI-MS using ribosomal subunit proteins as biomarkers. Anal. Chem. 79, 8712–8719 (2007).
Lartigue, M. F. et al. Identification of Streptococcus agalactiae isolates from various phylogenetic lineages by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 47, 2284–2287 (2009).
Mellmann, A. et al. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J. Clin. Microbiol. 46, 1946–1954 (2008).
Nagy, E., Maier, T., Urban, E., Terhes, G. & Kostrzewa, M. Species identification of clinical isolates of Bacteroides by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 15, 796–802 (2009).
Marklein, G. et al. Matrix-assisted laser desorption ionization-time of flight mass-spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47, 2912–2917 (2009).
Mullis, K. et al. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 51, 263–273 (1986).
Sauer, S. et al. Full flexibility genotyping of single nucleotide polymorphisms by the GOOD assay. Nucleic Acids Res. 28, e100 (2000).
Enright, M. C. & Spratt, B. G. Multilocus sequence typing. Trends Microbiol. 7, 482–487 (1999).
Nakamura, Y. et al. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science 235, 1616–1622 (1987).
Stanssens, P. et al. High-throughput MALDI-TOF discovery of genomic sequence polymorphisms. Genome Res. 14, 126–133 (2004). This paper describes the principle of MALDI-based resequencing.
Ehrich, M., Bocker, S. & van den Boom, D. Multiplexed discovery of sequence polymorphisms using base-specific cleavage and MALDI-TOF MS. Nucleic Acids Res. 33, e38 (2005).
Ehrich, M. et al. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc. Natl Acad. Sci. USA 102, 15785–15790 (2005).
von Wintzingerode, F. et al. Base-specific fragmentation of amplified 16S rRNA genes analyzed by mass spectrometry: a tool for rapid bacterial identification. Proc. Natl Acad. Sci. USA 99, 7039–7044 (2002).
Kirpekar, F. et al. Matrix assisted laser desorption/ionization mass spectrometry of enzymatically synthesized RNA up to 150 kDa. Nucleic Acids Res. 22, 3866–3870 (1994).
Honisch, C. et al. Automated comparative sequence analysis by base-specific cleavage and mass spectrometry for nucleic acid-based microbial typing. Proc. Natl Acad. Sci. USA 104, 10649–10654 (2007). This article describes in detail the application of MALDI resequencing for the analysis of bacteria.
Griebel, T., Brinkmeyer, M. & Bocker, S. EPoS: a modular software framework for phylogenetic analysis. Bioinformatics 24, 2399–2400 (2008).
Bocker, S. Simulating multiplexed SNP discovery rates using base-specific cleavage and mass spectrometry. Bioinformatics 23, e5–e11 (2007).
Bocker, S. SNP and mutation discovery using base-specific cleavage and MALDI-TOF mass spectrometry. Bioinformatics 19, i44–i53 (2003).
Blanc, D. S. The use of molecular typing for epidemiological surveillance and investigation of endemic nosocomial infections. Infect. Genet. Evol. 4, 193–197 (2004).
Tenover, F. C. et al. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Infect. Control Hosp. Epidemiol. 18, 426–439 (1997).
Honisch, C., Raghunathan, A., Cantor, C. R., Palsson, B. O. & van den Boom, D. High-throughput mutation detection underlying adaptive evolution of Escherichia coli-K12. Genome Res. 14, 2495–2502 (2004).
Sauer, S., Reinhardt, R., Lehrach, H. & Gut, I. G. Single-nucleotide polymorphisms: analysis by mass spectrometry. Nature Protoc. 1, 1761–1771 (2006).
Storm, N., Darnhofer-Patel, B., van den Boom, D. & Rodi, C. P. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol. Biol. 212, 241–262 (2003).
Ecker, D. J. et al. Ibis T5000: a universal biosensor approach for microbiology. Nature Rev. Microbiol. 6, 553–558 (2008).
Muddiman, D. C. et al. Characterization of PCR products from bacilli using electrospray ionization FTICR mass spectrometry. Anal. Chem. 68, 3705–3712 (1996). One of the first publications showing the high-resolution detection of a PCR product by ESI Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometry. Currently, a simple TOF analyser is used instead of the costly ICR device.
Wunschel, D. S. et al. Analysis of double-stranded polymerase chain reaction products from the Bacillus cereus group by electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry. Rapid Commun. Mass Spectrom. 10, 29–35 (1996).
Muddiman, D. C., Anderson, G. A., Hofstadler, S. A. & Smith, R. D. Length and base composition of PCR-amplified nucleic acids using mass measurements from electrospray ionization mass spectrometry. Anal. Chem. 69, 1543–1549 (1997).
Ecker, D. J. et al. Molecular genotyping of microbes by multilocus PCR and mass spectrometry: a new tool for hospital infection control and public health surveillance. Methods Mol. Biol. 551, 71–87 (2009). An informative introduction to the PCR–ESI detection method, including case studies.
Jiang, Y. & Hofstadler, S. A. A highly efficient and automated method of purifying and desalting PCR products for analysis by electrospray ionization mass spectrometry. Anal. Biochem. 316, 50–57 (2003).
Hofstadler, S. A. & Sannes-Lowery, K. A. Applications of ESI-MS in drug discovery: interrogation of noncovalent complexes. Nature Rev. Drug Discov. 5, 585–595 (2006).
Sauer, S. & Gut, I. G. Genotyping single-nucleotide polymorphisms by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 782, 73–87 (2002).
Hall, T. A. et al. Base composition analysis of human mitochondrial DNA using electrospray ionization mass spectrometry: a novel tool for the identification and differentiation of humans. Anal. Biochem. 344, 53–69 (2005).
Sampath, R. et al. Global surveillance of emerging influenza virus genotypes by mass spectrometry. PLoS ONE 2, e489 (2007).
Ecker, D. J. et al. Rapid identification and strain-typing of respiratory pathogens for epidemic surveillance. Proc. Natl Acad. Sci. USA 102, 8012–8017 (2005).
Ecker, J. A. et al. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J. Clin. Microbiol. 44, 2921–2932 (2006).
Van Ert, M. N. et al. Mass spectrometry provides accurate characterization of two genetic marker types in Bacillus anthracis. Biotechniques 37, 642–651 (2004).
Faix, D. J., Sherman, S. S. & Waterman, S. H. Rapid-test sensitivity for novel wwine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. (2009).
Hujer, K. M. et al. Rapid determination of quinolone resistance in Acinetobacter spp. J. Clin. Microbiol. 47, 1436–1442 (2009).
Keys, C. J. et al. Compilation of a MALDI-TOF mass spectral database for the rapid screening and characterisation of bacteria implicated in human infectious diseases. Infect. Genet. Evol. 4, 221–242 (2004).
Arnold, R. J. & Reilly, J. P. Fingerprint matching of E. coli strains with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry of whole cells using a modified correlation approach. Rapid Commun. Mass Spectrom. 12, 630–636 (1998).
Chen, P., Lu, Y. & Harrington, P. B. Application of linear and nonlinear discrete wavelet transforms to MALDI-MS measurements of bacteria for classification. Anal. Chem. 80, 7218–7225 (2008).
Hagen, R. M. et al. Development of a real-time PCR assay for rapid identification of methicillin-resistant Staphylococcus aureus from clinical samples. Int. J. Med. Microbiol. 295, 77–86 (2005).
He, Y. et al. PIML: the Pathogen Information Markup Language. Bioinformatics 21, 116–121 (2005).
Mellmann, A. et al. High interlaboratory reproducibility of matrix-assisted laser desorption ionization-time of flight mass spectrometry-based species identification of nonfermenting bacteria. J. Clin. Microbiol. 47, 3732–3734 (2009).
Guo, Z., Liu, Y., Li, S. & Yang, Z. Interaction of bacteria and ion-exchange particles and its potential in separation for matrix-assisted laser desorption/ionization mass spectrometric identification of bacteria in water. Rapid Commun. Mass Spectrom. 23, 3983–3993 (2009).
Acknowledgements
We thank M. Kostrzewa, T. Maier, J. Sauer and C. Honisch for discussions and C.-T. Han for critical reading of the manuscript. We also thank S. Holzhauser, V. Nicolaysen and A. Freiwald for help with the figures. Our work is supported by the German Ministry for Education and Research (BMBF; grant number 0315082), the National Genome Research Net (NGFN; grant number 01 GS 0828), the European Union (FP7/2007-2013, under grant agreement number [HEALTH-F4-2008-201418], entitled READNA), and the Max Planck Society. We apologize to all whose work is not covered here owing to space limitations.
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
Entrez Genome Project
Escherichia coli str. K-12 substr. MG1655
FURTHER INFORMATION
Glossary
- Enzyme-linked immunosorbent assay
-
(ELISA). A common serological test for the presence of particular antigens, using specific antibody binding and an enzyme to generate reporter label molecules.
- Pulsed-field gel electrophoresis
-
(PFGE). A variation on the standard gel electrophoresis such that an alternating voltage gradient is applied to improve the separation of biomolecules such as large nucleic acids.
- GC–MS
-
Gas chromatography–mass spectrometry coupling, a technique that is applied to isolate the components of volatile samples in the gas phase and to seamlessly identify and potentially quantify the single components by a mass spectrometer.
- Matrix-assisted laser desorption/ionization
-
(MALDI). A soft method for the generation of ions that uses laser bombardment of crystals containing analyte molecules and an excess of matrix molecules that absorb laser light of a specific wavelength.
- Electrospray ionization
-
(ESI). A soft technique for the generation of ions in aerosols.
- Unsupervised hierarchical clustering
-
A procedure for statistical data analysis that uses clustering algorithms to find successive data clusters using previously generated clusters, which can be represented in a tree structure termed a dendrogram.
- Multilocus sequence typing
-
A molecular biology method for the typing of multiple genomic loci, which is applied to analyse isolates of bacterial species using the DNA sequences of internal fragments of multiple housekeeping genes. The first protocol has been established for Neisseria meningitidis, the causative pathogen of meningococcal meningitis and septicaemia.
- Euclidean distance
-
A mathematical term that describes the distance between two points, which can be verified by application of the Pythagorean theorem.
Rights and permissions
About this article
Cite this article
Sauer, S., Kliem, M. Mass spectrometry tools for the classification and identification of bacteria. Nat Rev Microbiol 8, 74–82 (2010). https://doi.org/10.1038/nrmicro2243
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2243
This article is cited by
-
Mass spectrometry-based proteomics as an emerging tool in clinical laboratories
Clinical Proteomics (2023)
-
Long-term label-free assessments of individual bacteria using three-dimensional quantitative phase imaging and hydrogel-based immobilization
Scientific Reports (2023)
-
Efficient deep learning architectures for fast identification of bacterial strains in resource-constrained devices
Multimedia Tools and Applications (2022)
-
PhyloQuant approach provides insights into Trypanosoma cruzi evolution using a systems-wide mass spectrometry-based quantitative protein profile
Communications Biology (2021)
-
Assessing the ratio of Bacillus spores and vegetative cells by shotgun proteomics
Environmental Science and Pollution Research (2021)