Key Points
-
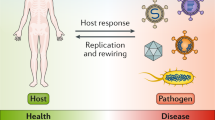
Study of infectious diseases involves a broad range of research disciplines from molecular biology to population dynamics. This article discusses the ways in which mathematical modelling can be used to integrate studies across these different disciplines.
-
The article focuses on tuberculosis (TB) as an example of a complex persistent infection with a major impact on global health.
-
Approaches to integrate molecular and cellular research with epidemiology are illustrated by a discussion of host and pathogenic diversity in the context of population-based models of TB.
-
Analogous population-based mathematical approaches can be used to model the immune response during TB.
-
An alternative approach of agent-based modelling is illustrated by a description of key factors that regulate formation of granulomas in TB.
Abstract
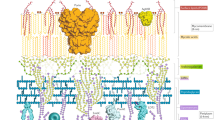
The human immune response does an excellent job of clearing most of the pathogens that we encounter throughout our lives. However, some pathogens persist for the lifetime of the host. Despite many years of research, scientists have yet to determine the basis of persistence of most pathogens, and have therefore struggled to develop reliable prevention and treatment strategies. Systems biology provides a new and integrative tool that will help to achieve these goals. In this article, we use Mycobacterium tuberculosis as an example of how systems-biology approaches have begun to make strides in uncovering important facets of the host–pathogen interaction.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Blaser, M. J. & Kirschner, D. The equilibria that allow bacterial persistence in human hosts. Nature 449, 843–849 (2007).
Kitano, H. Systems biology: a brief overview. Science 295, 1662–1664 (2002).
Yates, A., Chan, C. C., Callard, R. E., George, A. J. & Stark, J. An approach to modelling in immunology. Brief Bioinform. 2, 245–257 (2001).
Edelstein-Keshet, L. Mathematical Models in Biology (Random House, New York, 1988).
Murray, J. D. Mathematical Biology (Springer-Verlag, New York, 1989).
Keener, J. P. & Sneyd, J. Mathematical Physiology (Springer, New York, 1998).
Segel, L. A. & Cohen, I. R. Design Principles For the Immune System and Other Distributed Autonomous Systems (Oxford Univ. Press, 2001).
Grimm, V. & Railsback, S. F. Individual-Based Modeling and Ecology (Princeton Univ. Press, 2005).
Lauffenburger, D. A. & Linderman, J. L. Receptors: Models for Binding, Trafficking and Signaling (Oxford Univ. Press, 1993).
Brauer, F. & Castillo- Châavez, C. Mathematical Models in Population Biology and Epidemiology (Springer, New York, 2001).
Armitage, P., Berry. G. & Matthews, J. N. S. Statistical Methods in Medical Research (ed. Malden, M. A.) (Blackwell Science, Oxford, 2001).
Lund, O. Immunological Bioinformatics (The MIT Press, Cambridge, Massachusettes, 2005).
Ewens, W. J. & Grant, G. R. Statistical Methods in Bioinformatics: an Introduction (Springer, New York, 2005).
DeAngelis, D. L. & Gross, L. J. Individual-Based Models and Approaches in Ecology: Populations, Communities, and Ecosystems (Chapman & Hall, New York, 1992).
Stewart, G. R., Robertson, B. D. & Young, D. B. Tuberculosis: a problem with persistence. Nature Rev. Microbiol. 1, 97–105 (2003). Review of the biology of persistent infection with M. tuberculosis .
Fibonacci, L. & Sigler, L. E. Fibonacci's Liber Abaci: a Translation into Modern English of Leonardo Pisano's Book of Calculation (Springer, New York, 2002).
Dietz, K. & Heesterbeek, J. A. Bernoulli was ahead of modern epidemiology. Nature 408, 513–514 (2000).
Dietz, K. & Heesterbeek, J. A. Daniel Bernoulli's epidemiological model revisited. Math. Biosci. 180, 1–21 (2002).
Blower, S. & Bernoulli, D. An attempt at a new analysis of the mortality caused by smallpox and of the advantages of inoculation to prevent it. Rev. Med. Virol. 14, 275–288 (2004).
Farr, W. On the Cattle Plague. J. Soc. Sci. 1, 349–351 (1866).
Ross, R. An application of the theory of probabilities to the study of a priori pathometry. Part I. Proc. R. Soc. Lond. A 92, 204–230 (1916).
Ross, R. & Hudson, H. P. An application of the theory of probabilities to the study of a priori pathometry. Part II. Proc. R. Soc. Lond. B 89, 507 (1917).
Ross, R. & Hudson, H. P. An application of the theory of probabilities to the study of a priori pathometry. Part III. Proc. R. Soc. Lond. B 89, 507 (1917).
Kermack, W. O. & McKendrick, A. G. A contribution to the mathematical theory of epidemics. Proc. R. Soc. Lond. A 115, 700–721 (1927).
Kermack, W. O. & McKendrick, A. G. Contributions to the mathematical theory of epidemics. II. The problem of endemicity. Proc. R. Soc. Lond. A 138, 55–83 (1932).
Kermack, W. O. & McKendrick, A. G. Contributions to the mathematical theory of epidemics. III. Further studies of the problem of endemicity. Proc. R. Soc. Lond. A 141, 94–122 (1933).
Blower, S. M. et al. The intrinsic transmission dynamics of tuberculosis epidemics. Nature Med. 1, 815–821 (1995).
Murphy, B. M., Singer, B. H., Anderson, S. & Kirschner, D. Comparing epidemic tuberculosis in demographically distinct heterogeneous populations. Math. Biosci. 180, 161–185 (2002).
Waaler, H., Geser, A. & Andersen, S. The use of mathematical models in the study of the epidemiology of tuberculosis. Am. J. Public Health Nations Health 52, 1002–1013 (1962).
Blower, S. M., Small, P. M. & Hopewell, P. C. Control strategies for tuberculosis epidemics: new models for old problems. Science 273, 497–500 (1996). Developed a model for designing effective control strategies for TB that was used to assess how suboptimal programmes can contribute to the development of drug resistance.
Murphy, B. M., Singer, B. H. & Kirschner, D. On treatment of tuberculosis in heterogeneous populations. J. Theor. Biol. 223, 391–404 (2003).
Dye, C., Garnett, G. P., Sleeman, K. & Williams, B. G. Prospects for worldwide tuberculosis control under the WHO DOTS strategy. Directly observed short-course therapy. Lancet 352, 1886–1891 (1998).
Murray, C. J. & Salomon, J. A. Modeling the impact of global tuberculosis control strategies. Proc. Natl Acad. Sci. USA 95, 13881–13886 (1998).
Young, D. & Dye, C. The development and impact of tuberculosis vaccines. Cell 124, 683–687 (2006). Used epidemiology modelling to predict the impact of the combined effects of drug treatment and vaccination on TB control.
Dye, C. & Williams, B. G. Eliminating human tuberculosis in the twenty-first century. J. R. Soc. Interface 5, 653–662 (2008).
Keeling, M. J., Woolhouse, M. E., May, R. M., Davies, G. & Grenfell, B. T. Modelling vaccination strategies against foot-and-mouth disease. Nature 421, 136–142 (2003).
Keeling, M. J. et al. Dynamics of the 2001 UK foot and mouth epidemic: stochastic dispersal in a heterogeneous landscape. Science 294, 813–817 (2001).
Tildesley, M. J. et al. Optimal reactive vaccination strategies for a foot-and-mouth outbreak in the UK. Nature 440, 83–86 (2006).
Aparicio, J. P., Capurro, A. F. & Castillo-Chavez, C. Transmission and dynamics of tuberculosis on generalized households. J. Theor. Biol. 206, 327–341 (2000).
Colijn, C., Cohen, T. & Murray, M. Emergent heterogeneity in declining tuberculosis epidemics. J. Theor. Biol. 247, 765–774 (2007).
Grassly, N. C. & Fraser, C. Mathematical models of infectious disease transmission. Nature Rev. Microbiol. 6, 477–487 (2008).
Comstock, G. W. Tuberculosis in twins: a re-analysis of the Prophit survey. Am. Rev. Respir. Dis. 117, 621–624 (1978).
Alcais, A., Fieschi, C., Abel, L. & Casanova, J. L. Tuberculosis in children and adults: two distinct genetic diseases. J. Exp. Med. 202, 1617–1621 (2005).
Barreiro, L. B. et al. Promoter variation in the DC-SIGN-encoding gene CD209 is associated with tuberculosis. PLoS Med. 3, e20 (2006).
Bellamy, R. et al. Tuberculosis and chronic hepatitis B virus infection in Africans and variation in the vitamin D receptor gene. J. Infect. Dis. 179, 721–724 (1999).
Hoal-Van Helden, E. G. et al. Mannose-binding protein B allele confers protection against tuberculous meningitis. Pediatr. Res. 45, 459–464 (1999).
Khor, C. C. et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nature Genet. 39, 523–528 (2007).
Malik, S. et al. Alleles of the NRAMP1 gene are risk factors for pediatric tuberculosis disease. Proc. Natl Acad. Sci. USA 102, 12183–12188 (2005).
Marquet, S. & Schurr, E. Genetics of susceptibility to infectious diseases: tuberculosis and leprosy as examples. Drug Metab. Dispos. 29, 479–483 (2001).
Thuong, N. T. et al. A polymorphism in human TLR2 is associated with increased susceptibility to tuberculous meningitis. Genes Immun. 8, 422–428 (2007).
Van Soolingen, D. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Intern. Med. 249, 1–26 (2001).
Tanaka, M. M. & Rosenberg, N. A. Optimal estimation of transposition rates of insertion sequences for molecular epidemiology. Stat. Med. 20, 2409–2420 (2001).
Tanaka, M. M., Small, P. M., Salamon, H. & Feldman, M. W. The dynamics of repeated elements: applications to the epidemiology of tuberculosis. Proc. Natl Acad. Sci. USA 97, 3532–3537 (2000).
Gagneux, S. & Small, P. M. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect. Dis. 7, 328–337 (2007).
Gagneux, S. et al. Variable host–pathogen compatibility in Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA 103, 2869–2873 (2006).
Cohen, T. & Murray, M. Modeling epidemics of multidrug-resistant M. tuberculosis of heterogeneous fitness. Nature Med. 10, 1117–1121 (2004).
Kirschner, D. Dynamics of co-infection with M. tuberculosis and HIV-1. Theor. Popul. Biol. 55, 94–109 (1999).
Naresh, R. & Tripathi, A. Modelling and analysis of HIV–TB co-infection in a variable size population. Math. Model. Anal. 10, 275–286 (2005).
West, R. W. & Thompson, J. R. Modeling the impact of HIV on the spread of tuberculosis in the United States. Math. Biosci. 143, 35–60 (1997).
Porco, T. C., Small, P. M. & Blower, S. M. Amplification dynamics: predicting the effect of HIV on tuberculosis outbreaks. J. Acquir. Immune Defic. Syndr. 28, 437–444 (2001).
Marino, S. & Kirschner, D. E. The human immune response to Mycobacterium tuberculosis in lung and lymph node. J. Theor. Biol. 227, 463–486 (2004).
Marino, S. et al. Dendritic cell trafficking and antigen presentation in the human immune response to Mycobacterium tuberculosis. J. Immunol. 173, 494–506 (2004).
Megason, S. G. & Fraser, S. E. Imaging in systems biology. Cell 130, 784–795 (2007).
Cushing, J. M. An Introduction to Structured Population Dynamics (Society Industrial and Applied Mathematics, Philadelphia, 1998).
Yates, A., Bergmann, C., Van Hemmen, J. L., Stark, J. & Callard, R. Cytokine-modulated regulation of helper T cell populations. J. Theor. Biol. 206, 539–560 (2000).
Yates, A., Callard, R. & Stark, J. Combining cytokine signalling with T-bet and GATA-3 regulation in Th1 and Th2 differentiation: a model for cellular decision-making. J. Theor. Biol. 231, 181–196 (2004). Presented a modelling framework that showed how to integrate transcription-factor dynamics with cytokine signalling in a population of T cells.
Stark, J., Chan, C. & George, A. J. Oscillations in the immune system. Immunol. Rev. 216, 213–231 (2007).
Tyson, J. J., Chen, K. C. & Novak, B. Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Curr. Opin. Cell Biol. 15, 221–231 (2003).
Di Ventura, B., Lemerle, C., Michalodimitrakis, K. & Serrano, L. From in vivo to in silico biology and back. Nature 443, 527–533 (2006).
Kholodenko, B. N. Cell-signalling dynamics in time and space. Nature Rev. Mol. Cell Biol. 7, 165–176 (2006).
Brewer, D., Barenco, M., Callard, R., Hubank, M. & Stark, J. Fitting ordinary differential equations to short time course data. Philos. Transact. A Math. Phys. Eng. Sci. 366, 519–544 (2008).
Jaqaman, K. & Danuser, G. Linking data to models: data regression. Nature Rev. Mol. Cell Biol. 7, 813–819 (2006).
Kitano, H. Towards a theory of biological robustness. Mol. Syst. Biol. 3, 137 (2007).
Bailey, N. T. J. The Elements of Stochastic Processes with Applications to the Natural Sciences (Wiley, New York, 1964).
Ermentrout, G. B. & Edelstein-Keshet, L. Cellular automata approaches to biological modeling. J. Theor. Biol. 160, 97–133 (1993). The first study to use stochastic-type models.
Schnappinger, D. et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198, 693–704 (2003).
Pleissner, K. P. et al. Web-accessible proteome databases for microbial research. Proteomics 4, 1305–1313 (2004).
Sassetti, C. M., Boyd, D. H. & Rubin, E. J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 (2003).
Sassetti, C. M. & Rubin, E. J. Genetic requirements for mycobacterial survival during infection. Proc. Natl Acad. Sci. USA 100, 12989–12994 (2003).
Rengarajan, J., Bloom, B. R. & Rubin, E. J. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl Acad. Sci. USA 102, 8327–8332 (2005).
Stewart, G. R., Patel, J., Robertson, B. D., Rae, A. & Young, D. B. Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog. 1, 269–278 (2005).
Beste, D. J. et al. GSMN–TB: a web-based genome-scale network model of Mycobacterium tuberculosis metabolism. Genome Biol. 8, R89 (2007).
Jamshidi, N. & Palsson, B. O. Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst. Biol. 1, 26 (2007).
Hart, P. D., Armstrong, J. A., Brown, C. A. & Draper, P. Ultrastructural study of the behavior of macrophages toward parasitic mycobacteria. Infect. Immun. 5, 803–807 (1972).
Armstrong, J. A. & Hart, P. D. Phagosome–lysosome interactions in cultured macrophages infected with virulent tubercle bacilli. Reversal of the usual nonfusion pattern and observations on bacterial survival. J. Exp. Med. 142, 1–16 (1975).
Russell, D. G. Mycobacterium tuberculosis: here today, and here tomorrow. Nature Rev. Mol. Cell Biol. 2, 569–577 (2001).
van der Wel, N. et al. M. tuberculosis and M. leprae translocate from the phagolysosome to the cytosol in myeloid cells. Cell 129, 1287–1298 (2007).
Jordao, L., Bleck, C. K. E., Mayorga, L., Griffiths, G. & Anes, E. On the killing of mycobacteria by macrophages. Cell. Microbiol. 10, 529–548 (2008).
Ehrt, S. et al. Reprogramming of the macrophage transcriptome in response to interferon-γ and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 194, 1123–1140 (2001).
Gilchrist, M. et al. Systems biology approaches identify ATF3 as a negative regulator of Toll-like receptor 4. Nature 441, 173–178 (2006). One of the first successful attempts to apply systems-biology approaches to the analysis of the dynamic response of macrophages.
Roach, J. C. et al. Transcription factor expression in lipopolysaccharide-activated peripheral-blood-derived mononuclear cells. Proc. Natl Acad. Sci. USA 104, 16245–16250 (2007).
Reed, M. B. et al. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431, 84–87 (2004).
Newton, S. M. et al. A deletion defining a common Asian lineage of Mycobacterium tuberculosis associates with immune subversion. Proc. Natl Acad. Sci. USA 103, 15594–15598 (2006).
Chang, S., Linderman, J. & Kirschner, D. Multiple mechanisms allow Mycobacterium tuberculosis to continuously inhibit MHC class II-mediated antigen presentation by macrophages. Proc. Natl Acad. Sci. USA 102, 4530–4535 (2005).
Hund, T. J., Kucera, J. P., Otani, N. F. & Rudy, Y. Ionic charge conservation and long-term steady state in the Luo–Rudy dynamic cell model. Biophys. J. 81, 3324–3331 (2001).
Alarcon, T., Byrne, H. M. & Maini, P. K. Towards whole-organ modelling of tumour growth. Prog. Biophys. Mol. Biol. 85, 451–472 (2004). The first published paper to propose a multi-scale approach to understanding a host biological process.
Comisar, W. A., Hsiong, S. X., Kong, H. J., Mooney, D. J. & Linderman, J. J. Multi-scale modeling to predict ligand presentation within RGD nanopatterned hydrogels. Biomaterials 27, 2322–2329 (2006).
Kirschner, D. E., Chang, S. T., Riggs, T. W., Perry, N. & Linderman, J. J. Toward a multiscale model of antigen presentation in immunity. Immunol. Rev. 216, 93–118 (2007).
Kirschner, D. in In silico Immunology (eds. Flower, D. & Timmis, J.) 289–312 (Springer, New York, 2006).
Flynn, J. L. & Chan, J. Immunology of tuberculosis. Annu. Rev. Immunol. 19, 93–129 (2001).
Wigginton, J. E. & Kirschner, D. A model to predict cell-mediated immune regulatory mechanisms during human infection with Mycobacterium tuberculosis. J. Immunol. 166, 1951–1967 (2001). The first detailed mathematical model developed to study the host response to M. tuberculosis . Showed how virtual deletions and depletions can be used to perform studies that are not currently tractable in wet laboratories and predict mechanisms to explain experimental results.
Sud, D., Bigbee, C., Flynn, J. L. & Kirschner, D. E. Contribution of CD8+ T cells to control of Mycobacterium tuberculosis infection. J. Immunol. 176, 4296–4314 (2006).
Marino, S. et al. Differences in reactivation of tuberculosis induced from anti-TNF treatments are based on bioavailability in granulomatous tissue. PloS Comput. Biol. 3, e194 (2007).
Segovia-Juarez, J. L., Ganguli, S. & Kirschner, D. Identifying control mechanisms of granuloma formation during M. tuberculosis infection using an agent-based model. J. Theor. Biol. 231, 357–376 (2004). The first spatial, stochastic framework for studying granuloma formation in TB. This model predicted previously unidentified features of the immune response that are key to containing infection.
Marino, S., Hogue, I. B., Ray, C. J. & Kirschner, D. E. A methodology for performing global uncertainty and sensitivity analysis in systems biology. J. Theor. Biol. 20 Apr 2008 (doi:10.1016/j.jtbi.2008.04.011).
Acknowledgements
This Review was made possible by financial support from the UK Biotechnology and Biotechnology and Biological Sciences Research Council (BBSRC) via the Centre for Integrative Systems Biology at Imperial College (CISBIC), BB/C519670/1. Work described in this Review was supported, in part, by National Institutes of Health grants to D.K. (NIH R01 LM 009027 and NIH R01 HL 072682).
Author information
Authors and Affiliations
Corresponding author
Related links
Related links
DATABASES
Entrez Genome Project
FURTHER INFORMATION
Comprehensive Microbial Resource of the J. Craig Venter Institute
Pathogen website of the Wellcome Trust Sanger Institute
The Global Plan to Stop TB 2006–2015
Glossary
- Ordinary differential equation
-
A system of equations that is based on the rates (derivatives) of change of dependent variables with respect to time. Most of the interesting differential equations are nonlinear and, with a few exceptions, cannot be solved exactly. Approximate solutions are determined using computer simulations.
- Mendelian
-
Genetic inheritance of disease susceptibility through a single gene.
- TH1
-
After priming by exposure to signals from antigen-presenting cells, T cells undergo a process of maturation to their final effector phenotype. Cytokines produced by TH1 cells (for example, interferon-γ) enhance the antimicrobial activity of macrophages and have an important role in protection against Mycobacterium tuberculosis. Cytokines produced by TH2 cells (for example, interleukin-4) are important in promoting antibody responses. Cells that have not committed to the TH1 or TH2 lineages are referred to as TH0.
- Linkage analysis
-
A test for co-inheritance of genetic markers along with disease susceptibility in family groups.
- Cellular automata
-
Discrete models that consist of a regular grid of cells, each of which has a finite number of states. The state of a cell at time t is a function of the states of a finite number of cells (called its neighbourhood) at time t−1. Every cell has the same rule for updating that is based on the values in this neighbourhood. Each time the rules are applied to the whole grid, a new generation is created.
- Granuloma
-
A roughly spherical structure that comprises a focus of infection that is surrounded by immune cells. Dead cells at the centre of the granuloma may decompose, leaving a 'cheesy' residue that is referred to as caseum.
- Markov chain
-
A discrete-time stochastic process with the property that the next state solely depends on the present state, but not on the previous states. If a sequence of states has the Markov property, then every future state is conditionally independent of every prior state.
Rights and permissions
About this article
Cite this article
Young, D., Stark, J. & Kirschner, D. Systems biology of persistent infection: tuberculosis as a case study. Nat Rev Microbiol 6, 520–528 (2008). https://doi.org/10.1038/nrmicro1919
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1919
This article is cited by
-
A pursuit of Staphylococcus aureus continues: a role of persister cells
Archives of Pharmacal Research (2020)
-
Mycobacterium tuberculosis thymidylate kinase antigen assays for designating incipient, high-risk latent M.tb infection
BMC Infectious Diseases (2018)
-
Measuring the contributions of Chinese scholars to the research field of systems biology from 2005 to 2013
Scientometrics (2017)
-
Tuberculosis exposure, infection and disease in children: a systematic diagnostic approach
Pneumonia (2016)
-
Identification of the likely translational start of Mycobacterium tuberculosis GyrB
BMC Research Notes (2013)