Key Points
-
Over the past 20 years, molecular approaches, such as ribosomal RNA gene-sequence comparisons, have revolutionized our understanding of microbial diversity and ecology. Similarly, advances in genome sequencing technologies are now beginning to have a major impact on microbial ecology and the environmental sciences, particularly ocean science.
-
Several whole genome sequences for marine microorganisms have been completed and many more marine bacterial, archaeal and protistan genome sequencing projects are underway.
-
In addition to 'simple' genome sequencing, new genomic approaches to analyse natural microbial assemblages have been, and continue to be, developed. These new approaches, which are referred to by various names, including environmental genomics and metagenomics (the subject of this Focus issue), focus on cultivation-independent genomic survey strategies, such as the construction of large-insert bacterial artificial chromosome and fosmid libraries for large DNA inserts and the use of whole-genome shotgun sequencing for small DNA inserts.
-
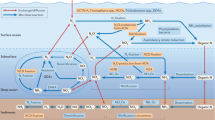
Three main case studies are discussed in detail: the discovery of bacterial proteorhodopsin using a phylogenetically anchored chromosome walking strategy; the shotgun sequence analysis of the microbial community in the Sargasso Sea; and the reconstruction of methane oxidation pathways in deep-sea archaea by shotgun sequencing of methanotroph-enriched fractions.
-
In the future, comparative community genomic approaches will hopefully begin to yield important, detailed information on microbial distribution, population structure and dynamics in various different environments.
Abstract
Marine microbial communities were among the first microbial communities to be studied using cultivation-independent genomic approaches. Ocean-going genomic studies are now providing a more comprehensive description of the organisms and processes that shape microbial community structure, function and dynamics in the sea. Through the lens of microbial community genomics, a more comprehensive view of uncultivated microbial species, gene and biochemical pathway distributions, and naturally occurring genomic variability is being brought into sharper focus. Besides providing new perspectives on oceanic microbial communities, these new studies are now poised to reveal the fundamental principles that drive microbial ecological and evolutionary processes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Woese, C. R. Bacterial evolution. Microbiol. Rev. 51, 221–271 (1987).
Olsen, G. J., Lane, D. J., Giovannoni, S. J., Pace, N. R. & Stahl, D. A. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40, 337–365 (1986).
Pace, N. R. A molecular view of microbial diversity and the biosphere. Science 276, 734–740 (1997).
Rappe, M. S. & Giovannoni, S. J. The uncultured microbial majority. Annu. Rev. Microbiol. 57, 369–394 (2003).
Hugenholtz, P., Goebel, B. M. & Pace, N. R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180, 4765–4774 (1998).
DeLong, E. F. Microbial seascapes revisited. Curr. Opin. Microbiol. 4, 290–295 (2001).
Wright, T. D., Vergin, K. L., Boyd, P. W. & Giovannoni, S. J. A novel δ-subdivision proteobacterial lineage from the lower ocean surface layer. Appl. Environ. Microbiol. 63, 1441–1448 (1997).
DeLong, E. F. Archaea in coastal marine environments. Proc. Natl Acad. Sci. USA 89, 5685–5689 (1992).
Bult, C. J. et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273, 1058–1073 (1996).
Dufresne, A. et al. Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome. Proc. Natl Acad. Sci. USA 100, 10020–10025 (2003).
Rocap, G. et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature 424, 1042–1047 (2003). This paper reports on the comparative genomic analysis of one of the most abundant photosynthetic organisms in the oceans. The authors report on depth-specific adaptations and genomic differences to gradients of light and nutrients found in the water column.
Palenik, B. et al. The genome of a motile marine Synechococcus. Nature 424, 1037–1042 (2003).
Hou, S. et al. Genome sequence of the deep-sea γ-proteobacterium Idiomarina loihiensis reveals amino acid fermentation as a source of carbon and energy. Proc. Natl Acad. Sci. USA 101, 18036–18041 (2004).
Moran, M. A. et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature 432, 910–913 (2004). This paper reports on the genome sequence of a marine bacterium related to Roseobacter spp., an α-proteobacterial group that is abundant in marine surface waters. The authors suggest the presence of specific adaptations to the marine environment with respect to carbon and sulphur metabolism, and test these hypotheses in physiological experiments.
Chen, C. Y. et al. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13, 2577–2587 (2003).
Makino, K. et al. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361, 743–749 (2003).
Nelson, K. E. et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature 399, 323–329 (1999).
Rabus, R. et al. The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ. Microbiol. 6, 887–902 (2004).
Glockner, F. O. et al. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc. Natl Acad. Sci. USA 100, 8298–8303 (2003).
Takami, H., Takaki, Y. & Uchiyama, I. Genome sequence of Oceanobacillus iheyensis isolated from the Iheya Ridge and its unexpected adaptive capabilities to extreme environments. Nucleic Acids Res. 30, 3927–3935 (2002).
Vezzi, A. et al. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307, 1459–1461 (2005). This paper reports the first DNA sequence of a piezophilic bacterium adapted to high hydrostatic pressures encountered in the deep sea. The sequence was used in microarray analyses to infer regulatory responses to changing hydrostatic pressure.
Ruby, E. G. et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl Acad. Sci. USA 102, 3004–3009 (2005).
Chinen, A., Uchiyama, I. & Kobayashi, I. Comparison between Pyrococcus horikoshii and Pyrococcus abyssi genome sequences reveals linkage of restriction–modification genes with large genome polymorphisms. Gene 259, 109–121 (2000).
Galagan, J. E. et al. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12, 532–542 (2002).
Hendrickson, E. L. et al. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 186, 6956–6969 (2004).
Kawarabayasi, Y. et al. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5, 55–76 (1998).
Kawarabayasi, Y. et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6, 83–101, 145–152 (1999).
Klenk, H. P. et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390, 364–370 (1997).
Robb, F. T. et al. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 330, 134–157 (2001).
Slesarev, A. I. et al. The complete genome of hyperthermophile Methanopyrus kandleri AV19 and monophyly of archaeal methanogens. Proc. Natl Acad. Sci. USA 99, 4644–4649 (2002).
Armbrust, E. V. et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86 (2004). This report describes the draft sequencing of the 34-Mb genome of marine diatom Thalassiosira pseudonana , which has 24 diploid nuclear chromosomes. Metabolic features inferred from the genome sequence were related to the growth strategies and ecology of this diatom. Evidence for the secondary endosymbioses with a red algal endosymbiont was found in the nuclear genome.
Schmidt, T. M., DeLong, E. F. & Pace, N. R. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J. Bacteriol. 173, 4371–4378 (1991).
Stein, J. L., Marsh, T. L., Wu, K. Y., Shizuya, H. & DeLong, E. F. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J. Bacteriol. 178, 591–599 (1996).
Béjà, O. et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289, 1902–1906 (2000).
Béjà, O. et al. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415, 630–633 (2002).
Breitbart, M. et al. Genomic analysis of uncultured marine viral communities. Proc. Natl Acad. Sci. USA 99, 14250–14255 (2002).
Breitbart, M. et al. Diversity and population structure of a near-shore marine-sediment viral community. Proc. Biol. Sci. 271, 565–574 (2004). This report describes the genetic make-up of two viral shotgun libraries from phage populations collected in coastal waters of San Diego. The authors show that double stranded DNA tailed phages and algal phages were well represented in the sample. Additionally, the most abundant viral group detected by shotgun cloning could represent upwards of 3% of the total phage.
de la Torre, J. R. et al. Proteorhodopsin genes are distributed among divergent marine bacterial taxa. Proc. Natl Acad. Sci. USA 100, 12830–12835 (2003).
Sabehi, G., Beja, O., Suzuki, M. T., Preston, C. M. & DeLong, E. F. Different SAR86 subgroups harbour divergent proteorhodopsins. Environ. Microbiol. 6, 903–910 (2004).
Schleper, C. et al. Genomic analysis reveals chromosomal variation in natural populations of the uncultured psychrophilic archaeon Cenarchaeum symbiosum. J. Bacteriol. 180, 5003–5009 (1998).
Vergin, K. L. et al. Screening of a fosmid library of marine environmental genomic DNA fragments reveals four clones related to members of the order Planctomycetales. Appl. Environ. Microbiol. 64, 3075–3078 (1998).
Rondon, M. R. et al. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66, 2541–2547 (2000).
Lopez-Garcia, P., Brochier, C., Moreira, D. & Rodriguez-Valera, F. Comparative analysis of a genome fragment of an uncultivated mesopelagic crenarchaeote reveals multiple horizontal gene transfers. Environ. Microbiol. 6, 19–34 (2004).
Treusch, A. H. et al. Characterization of large-insert DNA libraries from soil for environmental genomic studies of Archaea. Environ. Microbiol. 6, 970–980 (2004).
Moreira, D., Rodriguez-Valera, F. & Lopez-Garcia, P. Analysis of a genome fragment of a deep-sea uncultivated group II euryarchaeote containing 16S rDNA, a spectinomycin-like operon and several energy metabolism genes. Environ. Microbiol. 6, 959–969 (2004).
Quaiser, A. et al. Acidobacteria form a coherent but highly diverse group within the bacterial domain: evidence from environmental genomics. Mol. Microbiol. 50, 563–575 (2003).
Quaiser, A. et al. First insight into the genome of an uncultivated crenarchaeote from soil. Environ. Microbiol. 4, 603–611 (2002).
Schleper, C., Swanson, R. V., Mathur, E. J. & DeLong, E. F. Characterization of a DNA polymerase from the uncultivated psychrophilic archaeon Cenarchaeum symbiosum. J. Bacteriol. 179, 7803–7811 (1997).
Preston, C. M., Wu, K. Y., Molinski, T. F. & DeLong, E. F. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc. Natl Acad. Sci. USA 93, 6241–6246 (1996).
Handelsman, J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68, 669–685 (2004).
Riesenfeld, C. S., Schloss, P. D. & Handelsman, J. Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 38, 525–552 (2004).
Stein, J. L., Haygood, M. & Felbeck, H. Nucleotide sequence and expression of a deep-sea ribulose-1,5-bisphosphate carboxylase gene cloned from a chemoautotrophic bacterial endosymbiont. Proc. Natl Acad. Sci. USA 87, 8850–8854 (1990).
Cottrell, M. T., Moore, J. A. & Kirchman, D. L. Chitinases from uncultured marine microorganisms. Appl. Environ. Microbiol. 65, 2553–2557 (1999).
Hallam, S. J. et al. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 305, 1457–1462 (2004).
Tringe, S. G. et al. Comparative metagenomics of microbial communities. Science 308, 554–557 (2005).
DeLong, E. F. in Microbial Genomics (eds Fraser, C. M., Nelson., K. E. & Read, T. D.) 419–442 (Human Press Inc., Totowa, New Jersey, 2004).
DeLong, E. F. Microbial population genomics and ecology. Curr. Opin. Microbiol. 5, 520–524 (2002).
DeLong, E. F. Towards microbial systems science: integrating microbial perspective, from genomes to biomes. Environ. Microbiol. 4, 9–10 (2002).
Rodriguez-Valera, F. Approaches to prokaryotic biodiversity: a population genetics perspective. Environ. Microbiol. 4, 628–633 (2002).
Rodriguez-Valera, F. Environmental genomics, the big picture? FEMS Microbiol. Lett. 231, 153–158 (2004).
Beja, O. To BAC or not to BAC: marine ecogenomics. Curr. Opin. Biotechnol. 15, 187–190 (2004).
Doney, S. C., Abbott, M. R., Cullen, J. J., Karl, D. M. & Rothstein, L. From genes to ecosystems: the ocean's new frontier. Frontiers in Ecology and the Environment 2, 457–466 (2004).
Kim, U. -J., Shizuya, H., Dejong, P., Birren, B. & Simon, M. Stable propagation of cosmid sized human DNA inserts in an F-factor based vector. Nucleic Acids Res. 20, 1083–1185 (1992).
Shizuya, H. et al. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc. Natl Acad. Sci. USA 89, 8794–8797 (1992).
Béjà, O. et al. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2, 516–529 (2000).
Fleischmann, R. D. et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269, 496–512 (1995).
Tamas, I. et al. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296, 2376–2379 (2002).
Shigenobu, S., Watanabe, H., Hattori, M., Sakaki, Y. & Ishikawa, H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407, 81–86 (2000).
Tyson, G. W. et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428, 37–43 (2004).
Waterston, R. H., Lander, E. S. & Sulston, J. E. On the sequencing of the human genome. Proc. Natl Acad. Sci. USA 99, 3712–3716 (2002).
Waterston, R. H., Lander, E. S. & Sulston, J. E. More on the sequencing of the human genome. Proc. Natl Acad. Sci. USA 100, 3022–3024; author reply 3025–3026 (2003).
Oz, A., Sabehi, G., Koblizek, M., Massana, R. & Beja, O. Roseobacter-like bacteria in Red and Mediterranean Sea aerobic anoxygenic photosynthetic populations. Appl. Environ. Microbiol. 71, 344–353 (2005).
Sabehi, G. et al. Novel proteorhodopsin variants from the Mediterranean and Red Seas. Environ. Microbiol. 5, 842–849 (2003).
Man, D. et al. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 22, 1725–1731 (2003).
Béjà, O., Spudich, E. N., Spudich, J. L., Leclerc, M. & DeLong, E. F. Proteorhodopsin phototrophy in the ocean. Nature 411, 786–789 (2001).
Falkowski, P. G. & de Vargas, C. Shotgun sequencing in the sea: a blast from the past? Science 304, 58–60 (2004).
Venter, J. C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74 (2004).
Kruger, M. et al. A conspicuous nickel protein in microbial mats that oxidize methane anaerobically. Nature 426, 878–881 (2003).
Teeling, H., Meyerdierks, A., Bauer, M., Amann, R. & Glockner, F. O. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ. Microbiol. 6, 938–947 (2004).
Bielawski, J. P., Dunn, K. A., Sabehi, G. & Beja, O. Darwinian adaptation of proteorhodopsin to different light intensities in the marine environment. Proc. Natl Acad. Sci. USA 101, 14824–14829 (2004).
Krebs, R. A., Alexiev, U., Partha, R., DeVita, A. M. & Braiman, M. S. Detection of fast light-activated H+ release and M intermediate formation from proteorhodopsin. BMC Physiol. 2, 5 (2002).
Friedrich, T. et al. Proteorhodopsin is a light-driven proton pump with variable vectoriality. J. Mol. Biol. 321, 821–838 (2002).
Varo, G., Brown, L. S., Lakatos, M. & Lanyi, J. K. Characterization of the photochemical reaction cycle of proteorhodopsin. Biophys. J. 84, 1202–1207 (2003).
Lakatos, M., Lanyi, J. K., Szakacs, J. & Varo, G. The photochemical reaction cycle of proteorhodopsin at low pH. Biophys. J. 84, 3252–3256 (2003).
Dioumaev, A. K. et al. Proton transfers in the photochemical reaction cycle of proteorhodopsin. Biochemistry 41, 5348–5358 (2002).
Wang, W. W., Sineshchekov, O. A., Spudich, E. N. & Spudich, J. L. Spectroscopic and photochemical characterization of a deep ocean proteorhodopsin. J. Biol. Chem. 278, 33985–33991 (2003).
Lakatos, M. & Varo, G. The influence of water on the photochemical reaction cycle of proteorhodopsin at low and high pH. J. Photochem. Photobiol. B 73, 177–182 (2004).
Man-Aharonovich, D. et al. Characterization of RS29, a blue-green proteorhodopsin variant from the Red Sea. Photochem. Photobiol. Sci. 3, 459–462 (2004).
Bergo, V., Amsden, J. J., Spudich, E. N., Spudich, J. L. & Rothschild, K. J. Structural changes in the photoactive site of proteorhodopsin during the primary photoreaction. Biochemistry 43, 9075–9083 (2004).
Imasheva, E. S., Balashov, S. P., Wang, J. M., Dioumaev, A. K. & Lanyi, J. K. Selectivity of retinal photoisomerization in proteorhodopsin is controlled by aspartic acid 227. Biochemistry 43, 1648–1655 (2004).
Dioumaev, A. K., Wang, J. M., Balint, Z., Varo, G. & Lanyi, J. K. Proton transport by proteorhodopsin requires that the retinal Schiff base counterion Asp-97 be anionic. Biochemistry 42, 6582–6587 (2003).
Kelemen, B. R., Du, M. & Jensen, R. B. Proteorhodopsin in living color: diversity of spectral properties within living bacterial cells. Biochim. Biophys. Acta 1618, 25–32 (2003).
Kolber, Z. S., Van Dover, C. L., Niederman, R. A. & Falkowski, P. G. Bacterial photosynthesis in surface waters of the open ocean. Nature 407, 177–179 (2000).
Kolber, Z. S. et al. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science 292, 2492–2495 (2001).
Pradella, S. et al. Genome organization and localization of the pufLM genes of the photosynthesis reaction center in phylogenetically diverse marine α-proteobacteria. Appl. Environ. Microbiol. 70, 3360–3369 (2004).
Allgaier, M., Uphoff, H., Felske, A. & Wagner-Dobler, I. Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Appl. Environ. Microbiol. 69, 5051–5059 (2003).
Giovannoni, S. J., Britschgi, T. B., Moyer, C. L. & Field, K. G. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345, 60–63 (1990).
Acinas, S. G. et al. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430, 551–554 (2004).
Moore, L. R., Rocap, G. & Chisholm, S. W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393, 464–467 (1998).
Field, K. G. et al. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl. Environ. Microbiol. 63, 63–70 (1997).
Béjà, O. et al. Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Appl. Environ. Microbiol. 68, 335–345 (2002).
Garcia-Martinez, J. & Rodriguez-Valera, F. Microdiversity of uncultured marine prokaryotes: the SAR11 cluster and the marine Archaea of Group I. Mol. Ecol. 9, 935–948 (2000).
Klepac-Ceraj, V. et al. High overall diversity and dominance of microdiverse relationships in salt marsh sulphate-reducing bacteria. Environ. Microbiol. 6, 686–698 (2004).
Thompson, J. R. et al. Genotypic diversity within a natural coastal bacterioplankton population. Science 307, 1311–1313 (2005). This report examined the diversity of Vibrio splendidus isolates using rRNA and Hsp60 to examine the genetics of a single naturally occurring bacterial species. The extent of sympatric, co-occurring genetic diversity was remarkable, with this Vibrio species showing extensive allelic and genome size variation.
Acinas, S. G., Marcelino, L. A., Klepac-Ceraj, V. & Polz, M. F. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rrn operons. J. Bacteriol. 186, 2629–2635 (2004).
Hinrichs, K. U., Hayes, J. M., Sylva, S. P., Brewer, P. G. & DeLong, E. F. Methane-consuming archaebacteria in marine sediments. Nature 398, 802–805 (1999).
Orphan, V. J., House, C. H., Hinrichs, K. U., McKeegan, K. D. & DeLong, E. F. Multiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc. Natl Acad. Sci. USA 99, 7663–7668 (2002).
Orphan, V. J., House, C. H., Hinrichs, K. U., McKeegan, K. D. & DeLong, E. F. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293, 484–487 (2001).
Hallam, S. J., Girguis, P. R., Preston, C. M., Richardson, P. M. & DeLong, E. F. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl. Environ. Microbiol. 69, 5483–5491 (2003).
Detter, J. C. et al. Isothermal strand-displacement amplification applications for high-throughput genomics. Genomics 80, 691–698 (2002).
Suzuki, M. T. et al. Phylogenetic screening of ribosomal RNA gene-containing clones in bacterial artificial chromosome (BAC) libraries from different depths in Monterey Bay. Microb. Ecol. 48, 473–488 (2004).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Fitz-Gibbon, S. T. et al. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl Acad. Sci. USA 99, 984–989 (2002).
Acknowledgements
I am indebted to all my students and collaborators, past and present, for their dedication, inspiration and perspiration. Thanks to S. Hallam for providing Fig. 3 and to my oceanographic collaborators and colleagues, especially F. Chavez at the Monterey Bay Aquarium Research Institute and D. Karl at the University of Hawaii, for ongoing collaborative field efforts. The author's work is supported by the National Science Foundation, the Gordon and Betty Moore Foundation, and sequencing support from the Department of Energy (DoE) carried out at the DoE Joint Genome Institute.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Related links
DATABASES
Entrez
FURTHER INFORMATION
The Gordon and Betty Moore Foundation
NSF Microbial Sequencing Program FY 2005
The organism bins assembled from the Sargasso Sea WGS environmental sample dataset
Glossary
- EUPHOTIC ZONE
-
The uppermost stratum of the water column that receives sufficient light for photosynthesis.
- BENTHIC
-
Living in, or on the bottom of, a body of water.
- SPECIES RICHNESS
-
The number of different species in a given habitat, biotope, community or assemblage.
- SYMPATRIC
-
Populations, species or taxa occurring in the same geographical area.
- SPECIES EVENNESS
-
The relative abundance of species in a given habitat, biotope, community or assemblage.
- ALLOCHTHONOUS
-
Non-native. Found somewhere other than the place of origin.
- CONTINENTAL MARGIN
-
Shallow submarine extension of the continents, generally tens of meters deep, that extends seaward to the continental slope and the deep ocean.
Rights and permissions
About this article
Cite this article
DeLong, E. Microbial community genomics in the ocean. Nat Rev Microbiol 3, 459–469 (2005). https://doi.org/10.1038/nrmicro1158
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1158
This article is cited by
-
Himalayan Microbiomes for Agro-environmental Sustainability: Current Perspectives and Future Challenges
Microbial Ecology (2022)
-
Metalign: efficient alignment-based metagenomic profiling via containment min hash
Genome Biology (2020)
-
Temporal and spatial variations in the bacterial community composition in Lake Bosten, a large, brackish lake in China
Scientific Reports (2020)
-
Rapid screening of marine bacterial symbionts using MALDI-TOF MS
Archives of Microbiology (2020)