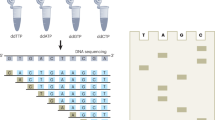
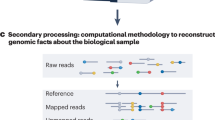
Key Points
-
Several academic and commercial research groups are working to develop new ultra-low-cost sequencing (ULCS) technologies. These aim to reduce the cost of DNA sequencing by several orders of magnitude.
-
ULCS technology could potentially have an important impact on human health by enabling the sequencing of 'personal genomes' as a component of individualized health care.
-
Microelectrophoretic approaches borrow microfabrication techniques from the semiconductor industry to miniaturize and integrate the amplification, purification and electrophoretic sequencing of DNA.
-
Sequencing by hybridization involves highly parallel genomic resequencing. It is carried out by hybridizing target DNA to high-density microarrays that are designed to query the identity of individual bases.
-
Cyclic-array methods that operate on amplified templates include 'fluorescent in situ sequencing', Pyrosequencing and 'massively parallel signature sequencing'. Cyclic-array methods that aim to directly sequence single molecules are also under development.
-
Methods such as nanopore sequencing offer the prospect of non-cyclic, real-time, single-molecule sequencing.
-
The prospect of ULCS and personal genomes raises various important ethical, legal and social questions.
Abstract
Nearly three decades have passed since the invention of electrophoretic methods for DNA sequencing. The exponential growth in the cost-effectiveness of sequencing has been driven by automation and by numerous creative refinements of Sanger sequencing, rather than through the invention of entirely new methods. Various novel sequencing technologies are being developed, each aspiring to reduce costs to the point at which the genomes of individual humans could be sequenced as part of routine health care. Here, we review these technologies, and discuss the potential impact of such a 'personal genome project' on both the research community and on society.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Collins, F. S., Morgan, M. & Patrinos, A. The human genome project: lessons from large-scale biology. Science 300, 286–290 (2003). A retrospective analysis of the Human Genome Project that was written by the top-level management.
National Human Genome Research Institute. Revolutionary Genome Sequencing Technologies — The $1000 Genome [online], <http://grants1.nih.gov/grants/guide/rfa-files/RFA-HG-04-003.html> (2004).
National Human Genome Research Institute. Near-term Technology Development for Genome Sequencing [online], <http://grants.nih.gov/grants/guide/rfa-files/RFA-HG-04-002.html> (2004).
Joneitz, E. Personal genomes. Technol. Rev. 104, 30 (2001).
Pray, L. A cheap personal genome? The Scientist [online], <http://www.biomedcentral.com/news/20021004/04> (2002).
Pennisi, E. Gene researchers hunt bargains, fixer-uppers. Science 298, 735–736 (2002).
Salisbury, M. W. Fourteen sequencing innovations that could change the way you work. Genome Technol. 35, 40–47 (2003).
Carroll, S. B. Genetics and the making of Homo sapiens. Nature 422, 840–857 (2003).
Ureta-Vidal, A., Ettwiller, L. & Birney, E. Comparative genomics: genome-wide analysis in metazoan eukaryotes. Nature Rev. Genet. 4, 251–62 (2003).
National Center for Biotechnology Information. GenBank Growth [online], <http://www.ncbi.nlm.nih.gov/Genbank/genbankstats.html> (2003).
National Center for Biotechnology Information. NCBI Taxonomy Browser [online], <http://www.ncbi.nlm.nih.gov/Taxonomy/txstat.cgi?uncultured=hide&unspecified=hide&m=0> (2004).
Integrated Genomics Inc. Genomes OnLine Database [online], <http://wit.integratedgenomics.com/GOLD> (2004).
Venter, J. C. et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science 4 Mar 2004 (doi:10.1126/science.1093857).
Gibbs, R. A. et al. The International HapMap Project. Nature 426, 789–796 (2003).
Holtzman, N. A. & Marteau T. M. Will genetics revolutionize medicine? N. Engl. J. Med. 343, 141–144 (2000).
Vitkup, D., Sander, C. & Church, G. M. The amino-acid mutational spectrum of human genetic disease. Genome Biol. 4, R72 (2003).
Farooqi, I. S. et al. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N. Engl. J. Med. 348, 1085–1095 (2003).
Smirnova, I. et al. Assay of locus-specific genetic load implicates rare Toll-like receptor 4 mutations in meningococcal susceptibility. Proc. Natl Acad. Sci. USA 100, 6075–6080 (2003).
Kruglyak, L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nature Genet. 22, 139–144 (1999).
Merz J. F., McGee G. E. & Sankar P. 'Iceland Inc.'?: On the ethics of commercial population genomics. Soc. Sci. Med. 58, 1201–1209 (2004).
Rajagopalan, H., Nowak, M. A., Vogelstein, B. & Lengauer, C. The significance of unstable chromosomes in colorectal cancer. Nature Rev. Cancer 3, 695–701 (2003).
Hanahan, D. & Weinberg, R. A. The hallmarks of cancer. Cell 10, 51–70 (2000).
Braich, R. S., Chelyapov, N., Johnson, C., Rothemund, P. W. & Adleman, L. Solution of a 20-variable 3-SAT problem on a DNA computer. Science 296, 499–450 (2002).
Reif, J. H. Computing: Successes and challenges. Science 296, 478–479 (2002).
Organisation for Economic Cooperation and Development (OECD). Health Data: Total Expenditure on Health, per capita US$ PPP [online], <http://www.oecd.org/dataoecd/1/33/2957315.xls> (2003).
Keith, J. M. et al. Unlocking hidden genomic sequence. Nucleic Acids Res. 32, e35 (2004).
Emrich, C. A., Tian, H., Medintz, I. L. & Mathies, R. A. Microfabricated 384-lane capillary array electrophoresis bioanalyzer for ultrahigh-throughput genetic analysis. Anal. Chem. 74, 5076–5083 (2002).
Koutny, L. et al. Eight hundred-base sequencing in a microfabricated electrophoretic device. Anal. Chem. 72, 3388–3391 (2000).
Paegel, B. M., Blazej, R. G. & Mathies, R. A. Microfluidic devices for DNA sequencing: sample preparation and electrophoretic analysis. Curr. Opin. Biotechnol. 14, 42–50 (2003). A review from the Mathies group on the integration of sample preparation and microelectrophoretic sequencing within a single microfluidic device.
Drmanac, S. Accurate sequencing by hybridization for DNA diagnostics and individual genomics. Nature Biotechnol. 16, 54–58 (1998).
Drmanac, R. et al. DNA sequencing by hybridization with arrays of samples or probes. Methods Mol. Biol. 170, 173–179 (2001).
Lipshutz, R. J. et al. Using oligonucleotide probe arrays to access genetic diversity. Biotechniques 19, 442–447 (1995).
Patil, N. et al. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294, 1719–1723 (2001). Perlegen's discovery of SNPs and haplotypes on human chromosome 21 through sequencing by hybridization.
Kruglyak, L. & Nickerson, D. A. Variation is the spice of life. Nature Genet. 27, 234–236 (2001).
Reich, D. E., Gabriel, S. B. & Altshuler, D. Quality and completeness of SNP databases. Nature Genet. 33, 457–458 (2003).
Church, G. M. & Gilbert, W. Genomic sequencing. Proc. Natl Acad. Sci. USA 81, 1991–1995 (1984).
Nowak, R. Getting the bugs worked out. Science 267, 172–174 (1995).
Ronaghi, M. Pyrosequencing sheds light on DNA sequencing. Genome Res. 11, 3–11 (2001).
Gharizadeh, B., Nordstrom, T., Ahmadian, A., Ronaghi, M. & Nyren, P. Long-read Pyrosequencing using pure 2′-deoxyadenosine-5′-O′-(1-thiotriphosphate) Sp-isomer. Analyt. Biochem. 301, 82–90 (2002). Describes the relatively long reads (50–100 bases) obtained through improvements to the Pyrosequencing sequencing-by-synthesis method.
Mitra, R. D., Shendure, J., Olejnik, J., Olejnik, E. K. & Church, G. M. Fluorescent in situ sequencing on polymerase colonies. Analyt. Biochem. 320, 55–65 (2003). Introduces the cyclic-array sequencing-by-synthesis technology that is being developed in the Church and Mitra laboratories.
Brenner, S. et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nature Biotechnol. 18, 630–634 (2000). Describes the 'massively parallel signature sequencing' (MPSS) technology that was developed by Lynx Therapeutics and involves cyclic-array sequencing by serial digestions, ligations and hybridizations.
Leamon, J. H. et al. A massively parallel PicoTiterPlate based platform for discrete picoliter-scale polymerase chain reactions. Electrophoresis 24, 3769–2777 (2003).
Sarkis, G. et al. Sequence analysis of the pAdEasy-1 recombinant adenoviral construct using the 454 Life Sciences sequencing-by-synthesis method. NCBI AY370911, gi:34014919 (2003).
Mitra, R. D. & Church, G. M. In situ localized amplification and contact replication of many individual DNA molecules. Nucleic Acids Res. 27, e34 (1999).
Mitra, R. D. et al. Digital genotyping and haplotyping with polymerase colonies. Proc. Natl Acad. Sci. USA 100, 5926–5931 (2003).
Dressman, D., Yan, H., Traverso, G., Kinzler, K. W. & Vogelstein, B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl Acad. Sci. USA 100, 8817–8822 (2003).
Metzker, M. L., et al. Termination of DNA synthesis by novel 3′-modified-deoxyribonucleoside 5—triphosphates. Nucleic Acids Res. 22, 4259–4267 (1994).
Welch, M. & Burgess, K. Synthesis of fluorescent, photolabile 3′-O-protected nucleoside triphosphates for the base addition sequencing scheme. Nucleosides Nucleotides 18, 197–199 (1999).
Hennig, C. AnyBase.nucleotides. GenoVoxx [online], <http://www.genovoxx.De> (2004).
Braslavsky, I., Hebert, B., Kartalov, E. & Quake, S. R. Sequence information can be obtained from single DNA molecules. Proc. Natl Acad. Sci. USA 100, 3960–3964 (2003). A description of single-molecule cyclic-array sequencing.
Levene, M. J. et al. Zero-mode waveguides for single-molecule analysis at high concentrations. Science 299, 682–686 (2003). Describes the detection of single-nucleotide incorporation events in zeptolitre-scale observation volumes.
Dean, F. B. et al. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl Acad. Sci. USA 99, 5261–5266 (2002).
Nelson, J. R. et al. TempliPhi, phi29 DNA polymerase based rolling circle amplification of templates for DNA sequencing. Biotechniques (Suppl.), 44–47 (2002).
Sorensen, K. J., Turteltaub, K., Vrankovich, G., Williams, J. & Christian, A. T. Whole-genome amplification of DNA from residual cells left by incidental contact. Anal. Biochem. 324, 312–314 (2004).
Rook, M. S., Delach, S. M., Deyneko, G., Worlock, A. & Wolfe, J. L. Whole genome amplification of DNA from laser capture-microdissected tissue for high-throughput single nucleotide polymorphism and short tandem repeat genotyping. Am. J. Pathol. 164, 23–33 (2003).
Zhu, J., Shendure, J., Mitra, R. D. & Church, G. M. Single molecule profiling of alternative pre-mRNA splicing. Science 301, 836–838 (2003).
Deamer, D. W. & Branton, D. Characterization of nucleic acids by nanopore analysis. Acc. Chem. Res. 35, 817–825 (2002).
Li, J., Gershow, M., Stein, D., Brandin, E. & Golovchenko, J. A. DNA molecules and configurations in a solid-state nanopore microscope. Nature Mater. 2, 611–615 (2003).
Deamer, D. W. & Akeson, M. Nanopores and nucleic acids: prospects for ultrarapid sequencing. Trends Biotechnol. 18, 147–151 (2000). A consideration of the successes and remaining challenges of nanopore sequencing.
Winters-Hilt, S. et al. Accurate classification of basepairs on termini of single DNA molecules. Biophys. J. 84, 967–976 (2003).
Hardin, S. H. Technologies at VisiGen. VisiGen Biotechnologies, Inc. [online], <http://www.visigenbio.com/tech.html> (2004).
Williams, J. Heterogenous assay for pyrophosphate. US Patent 6,306,607 (2001).
Robertson, J. A. The $1000 genome: ethical and legal issues in whole genome sequencing of individuals. Am. J. Bioeth. 3, W-IF1. (2003). A well-written overview of the ethical and legal implications of personal genomes.
Cooper, D. N. et al. Statistics. Human Gene Mutation Database Cardiff (HGMD) [online], <http://archive.uwcm.ac.uk/uwcm/mg/docs/hahaha.Html> (2004).
Oak Ridge National Laboratory. Genetics Privacy and Legislation. Human Genome Project Information [online], <http://www.ornl.gov/TechResources/Human_Genome/elsi/legislat.Html> (2003).
Gostin, L. O., Hodge, J. G. & Calvo, C. Genetics Policy & Law: A Report for Policymakers (National Council of State Legislators, Washington DC, 2001).
Biotechnological Process Patent Act. Pub. L. No. 104–41, 104th Cong., lst Sess. (1 Nov 1995).
Venter, J. C. A part of the human genome sequence. Science 299, 1183–1184 (2003).
Witelson, S. F., Kigar, D. L. & Harvey, T. The exceptional brain of Albert Einstein. Lancet 19, 2149–2153 (1999).
National Library of Medicine. The Visible Human Project [online], <http://www.nlm.nih.gov/research/visible/visible_human.Html> (2003).
U.S. Government. US Federal Government announcement of first Federal E-Gov Health Information Exchange Standards. egov Press Releases [online], <http://www.whitehouse.gov/omb/egov/press/chi_march.Htm> (2003).
Nurses' Health Study at Brigham and Women's Hospital [online], <http://www.nurseshealthstudy.Org> (2003).
Freimer, N. & Sabatti, C. The Human Phenome Project. Nature Genet. 3, 15–21 (2003).
Shendure, J. S., Mitra, R. D., Varma, C. & Church, G. M. Personal Genome Project [online], <http://arep.med.harvard.edu/PGP> (2004).
Public Library of Science (PLoS) [online], <http://www.plos.org/cgi-bin/plosSigned.pl> (2004).
SourceForge [online], <http://sourceforge.net> (2004).
The GNU Prjoect [online], <http://www.gnu.org> (2004).
Sanger, F., Nicklen, S. & Coulson, A. R. DNA sequencing with chain-terminating inhibitors. Proc. Natl Acad. Sci. USA 74, 5463–5467 (1977).
Maxam, A. M. & Gilbert, W. A new method for sequencing DNA. Proc. Natl Acad. Sci. USA 74, 560–564 (1977).
Gilbert, W. DNA sequencing and gene structure. Science 214, 1305–1312 (1981).
Sanger, F. Sequences, sequences, and sequences. Annu. Rev. Biochem. 5, 1–28 (1988).
Smith, L. M. et al. Fluorescence detection in automated DNA sequence analysis. Nature 321, 674–679 (1986).
Cook-Deegan, R. M. The Alta summit, December 1984. Genomics 5, 661–663 (1989).
Leder, P. Can the human genome project be saved from its critics ... and itself? Cell 63, 1–3 (1990).
Davis, B. D. The human genome and other initiatives. Science 249, 342–343 (1990).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Saha, S. et al. Using the transcriptome to annotate the genome. Nature Biotechnol. 20, 508–512 (2002).
Reymond, A. et al. Human chromosome 21 gene expression atlas in the mouse. Nature 420, 582–586 (2002).
Hahn, W. C. & Weinberg, R. A. Mechanisms of disease: rules for making human tumor cells. N. Engl. J. Med. 34, 1593–1603 (2002).
Paulson, T. G., Galipeau, P. C. & Reid, B. J. Loss of heterozygosity analysis using whole genome amplification, cell sorting, and fluorescence-based PCR. Genome Res. 9, 482–491 (1999).
Golub, T. R. et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286, 531–537 (1999).
Ramaswamy, S. et al. A molecular signature of metastasis in primary solid tumors. Nature Genet. 33, 49–54 (2003).
Weber, G., Shendure, J., Tanenbaum, D. M., Church, G. M., & Meyerson, M. Microbial sequence identification by computational subtraction of the human transcriptome. Nature Genet. 30, 141–142 (2002).
Stenger, D. A., Andreadis, J. D., Vora, G. J. & Pancrazio, J. J. Potential applications of DNA microarrays in biodefense-related diagnostics. Curr. Opin. Biotechnol. 13, 208–212 (2002).
Boffelli, D. et al. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science 299, 1391–1394 (2003).
Roberts, G. C. & Smith, C. W. Alternative splicing: combinatorial output from the genome. Curr. Opin. Chem. Biol. 6, 375–383 (2002).
Robyr, D. et al. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109, 437–446 (2002).
Yatabe, Y., Tavare, S. & Shibata, D. Investigating stem cells in human colon by using methylation patterns. Proc. Natl Acad. Sci. USA 9, 10839–10844 (2001).
Dymecki, S. M., Rodriguez, C. I. & Awatramani, R. B. Switching on lineage tracers using site-specific recombination. Methods Mol. Biol. 18, 309–334 (2002).
Lenski, R. E., Winkworth, C. L. & Riley, M. A. Rates of DNA sequence evolution in experimental populations of Escherichia coli during 20,000 generations. J. Mol. Evol. 56, 498–508 (2003).
Cooper, T. F., Rozen, D. E. & Lenski, R. E. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl Acad. Sci. USA 100, 1072–1077 (2003).
Gillespie, D. E. et al. Isolation of antibiotics turbomycin a and B from a metagenomic library of soil microbial DNA. Appl. Environ. Microbiol. 68, 4301–4306 (2002).
Badarinarayana, V. et al. Selection analyses of insertional mutants using subgenic-resolution arrays. Nature Biotechnol. 1, 1060–1065 (2001).
Sassetti, C. M., Boyd D. H. & Rubin E. J. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84 (2003).
Cerchia, L., Hamm, J., Libri, D., Tavitian, B. & de Franciscis, V. Nucleic acid aptamers in cancer medicine. FEBS Lett. 528, 12–16 (2002).
Cutler, D. J. et al. High-throughput variation detection and genotyping using microarrays. Genome Res. 11, 1913–1925 (2001).
Kurzweil, R. The 21st century: a confluence of accelerating revolutions. KurzweilAI.net [online], <http://www.kurzweilai.net/meme/frame.html?main=/articles/art0184.html> (2001).
Zakon, R. F. Hobbes' Internet Timeline [online], <http://www.zakon.org/robert/internet/timeline> (2004).
Acknowledgements
The authors thank members of the polony community and C. Hennig for sharing unpublished work, T. Wu and G. Porreca for helpful discussions, and R. Shendure and K. McKernan for critical reading of the manuscript.
Author information
Authors and Affiliations
Related links
Related links
DATABASES
LocusLink
OMIM
FURTHER INFORMATION
Revolutionary Genome Sequencing Technologies — The $1000 Genome
Glossary
- BERMUDA PRINCIPLES
-
A commitment that was made in Bermuda (February 1996) by an international assortment of genome-research sponsors to the principles of public sharing and the rapid release of human genome sequence information.
- 'COMMON' SINGLE NUCLEOTIDE POLYMORPHISMS
-
(SNPs). Those single-nucleotide substitutions that occur with an allelic frequency of more than 1% in a given population.
- HAPLOTYPE MAPPING
-
A technique that involves the use of combinations of 'common' DNA polymorphisms to find blocks of association with phenotypic traits.
- GENETIC HETEROGENEITY
-
Describes situations in which a similar phenotype can result from various genetic defects.
- SYNTHETIC BIOLOGY
-
A discipline that embraces the emerging ability to design, synthesize and evolve new genomes or biomimetic systems.
- PHYLOGENETIC FOOTPRINTING AND SHADOWING
-
The annotation of functional elements in a genome through bioinformatic comparisons to the genomes of one or more related species.
- DIRECTED EVOLUTION
-
The evolution of a protein (or organism) in the laboratory through rounds of mutation and selection for a particular activity or trait.
- FLUORESCENT IN SITU SEQUENCING (FISSEQ).
-
A cyclical, polymerase-driven sequencing method in which nucleotides are modified with fluorescent labels that can be chemically removed at each step.
- PHARMACOGENETIC
-
The heritable component of variation among individuals with respect to drug response or adverse reaction.
- SANGER SEQUENCING
-
(Chain termination or dideoxy method). A technique that uses an enzymatic procedure to synthesize DNA chains of varying length in four different reactions, stopping the DNA replication at positions that are occupied by one of the four bases, and then determining the resulting fragment lengths to decipher the sequence.
- POLONY
-
A colony of PCR amplicons that is derived from a single molecule of nucleic acid, amplified in situ in an acrylamide gel.
- TYPE IIS RESTRICTION ENZYME
-
A type of restriction endonuclease that is characterized by an asymmetric recognition site and cleavage at a fixed distance outside the recognition site.
- RAW READ
-
The actual nucleotide sequence that is generated by a sequencing instrument. This contrasts with the finished sequence, which is produced by obtaining the consensus sequence of many overlapping raw reads.
- FLUORESCENCE RESONANCE ENERGY TRANSFER
-
(FRET). A phenomenon by which excitation is transferred from a donor fluorescent molecule to an acceptor fluorescent molecule; the interaction is distance-dependent and can be used to probe molecular interactions at distances below the limit of optical resolution.
- ZERO-MODE WAVEGUIDE
-
A nanostructure device with physical properties that markedly limit the effective volume of observation.
- DEPHASING
-
The progressive loss of synchronization between templates within features as a consequence of the failure to achieve 100% extension at each extension cycle.
- MULTIPLE-DISPLACEMENT AMPLIFICATION
-
A technique for achieving whole-genome amplification that uses a strand-displacing polymerase to catalyse the isothermal (that is, at a constant temperature) amplification of DNA.
- WHOLE-GENOME AMPLIFICATION
-
(WGA). The in vitro amplification of a full genome sequence, ideally with even representation of the genome in the amplified product. Techniques for achieving WGA include PCR primed with random or degenerate oligonucleotides, or multiple-displacement amplification.
- COMPUTED TOMOGRAPHY
-
(CT). An imaging technology that uses computer processing of X-ray images to visualize cross-sectional (transverse) slices of internal structures; the advantage of CT over conventional radiography is the ability to eliminate superimposition.
- MAGNETIC RESONANCE [IMAGING]
-
(MRI). A non-invasive technique that is used to generate multi-dimensional proton-density images of internal organs, structures and lesions.
Rights and permissions
About this article
Cite this article
Shendure, J., Mitra, R., Varma, C. et al. Advanced sequencing technologies: methods and goals. Nat Rev Genet 5, 335–344 (2004). https://doi.org/10.1038/nrg1325
Issue Date:
DOI: https://doi.org/10.1038/nrg1325
This article is cited by
-
Molecular research progress and improvement approach of fruit quality traits in cucumber
Theoretical and Applied Genetics (2021)
-
Solid-State Nanopore
Nanoscale Research Letters (2018)
-
A genome-wide SNP-based genetic map and QTL mapping for agronomic traits in Chinese cabbage
Scientific Reports (2017)