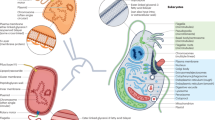
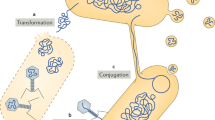
Key Points
-
Many horizontal gene transfer (HGT) events are initially neutral or nearly neutral (slightly detrimental) and can later adapt to become beneficial to the recipient.
-
HGT events often involve compound mobile genetic elements that promote their own dissemination by associating with adaptive traits in the gene pool of the mobilome.
-
HGT is most frequent between closely related species with highly similar genome features.
-
Large multicellular eukaryotes can evolve through gene acquisitions by the associated microbiome, which responds more quickly than the host to variation in environmental conditions.
-
Endosymbiotic gene transfer frequently involves gene import not only from the endosymbiotic donor to the host, but also from other organisms that contribute to the initial establishment of the endosymbiotic relationship.
-
Novel traits that evolve through HGT can lead to the exploitation of new niches, prompting an adaptive radiation to exploit the new resource without competition.
Abstract
Horizontal gene transfer (HGT) is the sharing of genetic material between organisms that are not in a parent–offspring relationship. HGT is a widely recognized mechanism for adaptation in bacteria and archaea. Microbial antibiotic resistance and pathogenicity are often associated with HGT, but the scope of HGT extends far beyond disease-causing organisms. In this Review, we describe how HGT has shaped the web of life using examples of HGT among prokaryotes, between prokaryotes and eukaryotes, and even between multicellular eukaryotes. We discuss replacement and additive HGT, the proposed mechanisms of HGT, selective forces that influence HGT, and the evolutionary impact of HGT on ancestral populations and existing populations such as the human microbiome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Tatum, E. L. & Lederberg, J. Gene recombination in the bacterium Escherichia coli. J. Bacteriol. 53, 673–684 (1947).
Went, F. W. Parallel evolution. Taxon 20, 197–226 (1971).
Treangen, T. J. & Rocha, E. P. C. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 7, e1001284 (2011).
Swithers, K. S., Soucy, S. M. & Gogarten, J. P. The role of reticulate evolution in creating innovation and complexity. Int. J. Evol. Biol. 2012, 418964 (2012).
Gogarten, J. P. & Townsend, J. P. Horizontal gene transfer, genome innovation and evolution. Nat. Rev. Microbiol. 3, 679–687 (2005).
Park, C. & Zhang, J. High expression hampers horizontal gene transfer. Genome Biol. Evol. 4, 523–532 (2012). This paper examines the impact of expression level on the transferability of a gene in both environmental and laboratory populations of E. coli.
Boto, L. Horizontal gene transfer in the acquisition of novel traits by metazoans. Proc. Biol. Sci. 281, 20132450 (2014).
Huang, J. Horizontal gene transfer in eukaryotes: the weak-link model. Bioessays 35, 868–875 (2013). This letter proposes a model for ongoing HGT in eukaryotes involving unicellular and early developmental stages to overcome the barrier of genome sequestration in eukaryotes.
Andersson, J. O. Gene transfer and diversification of microbial eukaryotes. Annu. Rev. Microbiol. 63, 177–193 (2009).
Timmis, J. N., Ayliffe, M. A., Huang, C. Y. & Martin, W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 (2004).
Koonin, E. V. Darwinian evolution in the light of genomics. Nucleic Acids Res. 37, 1011–1034 (2009).
Crisp, A., Boschetti, C., Perry, M., Tunnacliffe, A. & Micklem, G. Expression of multiple horizontally acquired genes is a hallmark of both vertebrate and invertebrate genomes. Genome Biol. 16, 50 (2015).
Riley, D. R. et al. Bacteria–human somatic cell lateral gene transfer is enriched in cancer samples. PLoS Comput. Biol. 9, e1003107 (2013).
Norman, A., Hansen, L. H. & Sørensen, S. J. Conjugative plasmids: vessels of the communal gene pool. Phil. Trans. R. Soc. B 364, 2275–2289 (2009).
Kyndt, T. et al. The genome of cultivated sweet potato contains Agrobacterium T-DNAs with expressed genes: an example of a naturally transgenic food crop. Proc. Natl Acad. Sci. USA 112, 201419685 (2015).
Johnston, C., Martin, B., Fichant, G., Polard, P. & Claverys, J.-P. Bacterial transformation: distribution, shared mechanisms and divergent control. Nat. Rev. Microbiol. 12, 181–196 (2014).
Chimileski, S., Dolas, K., Naor, A., Gophna, U. & Papke, R. T. Extracellular DNA metabolism in Haloferax volcanii. Front. Microbiol. 5, 57 (2014).
Lang, A. S., Zhaxybayeva, O. & Beatty, J. T. Gene transfer agents: phage-like elements of genetic exchange. Nat. Rev. Microbiol. 10, 472–482 (2012).
Naor, A. & Gophna, U. Cell fusion and hybrids in Archaea: prospects for genome shuffling and accelerated strain development for biotechnology. Bioengineered 4, 126–129 (2013).
Schleper, C., Holz, I., Janekovic, D., Murphy, J. & Zillig, W. A multicopy plasmid of the extremely thermophilic archaeon Sulfolobus effects its transfer to recipients by mating. J. Bacteriol. 177, 4417–4426 (1995).
Mevarech, M. & Werczberger, R. Genetic transfer in Halobacterium volcanii. J. Bacteriol. 162, 461–462 (1985).
Naor, A., Lapierre, P., Mevarech, M., Papke, R. T. & Gophna, U. Low species barriers in halophilic archaea and the formation of recombinant hybrids. Curr. Biol. 22, 1444–1448 (2012).
Dunning Hotopp, J. C. et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756 (2007). This paper describes HGT between Wolbachia spp., an intracellular bacterial symbiont, and its multicellular eukaryotic insect hosts.
Nikoh, N. et al. Bacterial genes in the aphid genome: absence of functional gene transfer from Buchnera to its host. PLoS Genet. 6, e1000827 (2010).
Moustafa, A. et al. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science 324, 1724–1726 (2009).
Chapman, J. A. et al. The dynamic genome of Hydra. Nature 464, 592–596 (2010).
Doolittle, W. F. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 14, 307–311 (1998).
Yue, J., Sun, G., Hu, X. & Huang, J. The scale and evolutionary significance of horizontal gene transfer in the choanoflagellate Monosiga brevicollis. BMC Genomics 14, 729 (2013).
Grant, J. R. & Katz, L. A. Phylogenomic study indicates widespread lateral gene transfer in entamoeba and suggests a past intimate relationship with parabasalids. Genome Biol. Evol. 6, 2350–2360 (2014).
Huang, J. & Gogarten, J. P. Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol. 8, R99 (2007). This paper discusses a complex tripartite relationship between a eukaryotic host, a cyanobacterium and a chlamydia that may have facilitated the establishment of modern plastids.
Graham, L. A., Li, J., Davidson, W. S. & Davies, P. L. Smelt was the likely beneficiary of an antifreeze gene laterally transferred between fishes. BMC Evol. Biol. 12, 190 (2012).
Yue, J., Hu, X., Sun, H., Yang, Y. & Huang, J. Widespread impact of horizontal gene transfer on plant colonization of land. Nat. Commun. 3, 1152 (2012).
Stewart, C. N., Halfhill, M. D. & Warwick, S. I. Transgene introgression from genetically modified crops to their wild relatives. Nat. Rev. Genet. 4, 806–817 (2003).
Evans, P. D., Mekel-Bobrov, N., Vallender, E. J., Hudson, R. R. & Lahn, B. T. Evidence that the adaptive allele of the brain size gene microcephalin introgressed into Homo sapiens from an archaic Homo lineage. Proc. Natl Acad. Sci. USA 103, 18178–18183 (2006).
Williams, D. et al. A rooted net of life. Biol. Direct 6, 45 (2011).
Gogarten, J. P., Doolittle, W. F. & Lawrence, J. G. Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 19, 2226–2238 (2002).
Colston, S. M. et al. Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case. mBio 5 e02136-14 (2014).
Delamuta, J. R. M., Ribeiro, R. A., Menna, P., Bangel, E. V. & Hungria, M. Multilocus sequence analysis (MLSA) of Bradyrhizobium strains: revealing high diversity of tropical diazotrophic symbiotic bacteria. Braz. J. Microbiol. 43, 698–710 (2012).
Williams, D., Gogarten, J. P. & Papke, R. T. Quantifying homologous replacement of loci between haloarchaeal species. Genome Biol. Evol. 4, 1223–1244 (2012).
Andam, C. P. & Gogarten, J. P. Biased gene transfer in microbial evolution. Nat. Rev. Microbiol. 9, 543–555 (2011).
Polz, M., Alm, E. & Hanage, W. Horizontal gene transfer and the evolution of bacterial. 29, 170–175 (2015). This paper investigates the interplay between HGT, population structure and lineage divergence in bacteria and archaea.
Langille, M. G. I., Hsiao, W. W. L. & Brinkman, F. S. L. Detecting genomic islands using bioinformatics approaches. Nat. Rev. Microbiol. 8, 373–382 (2010).
Daubin, V. & Ochman, H. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 14, 1036–1042 (2004).
Ragan, M. A. On surrogate methods for detecting lateral gene transfer. FEMS Microbiol. Lett. 201, 187–191 (2001).
Lukjancenko, O., Wassenaar, T. M. & Ussery, D. W. Comparison of 61 sequenced Escherichia coli genomes. Microb. Ecol. 60, 708–720 (2010).
Tettelin, H. et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial 'pan-genome'. Proc. Natl Acad. Sci. USA 102, 13950–13955 (2005).
Read, B. A. et al. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499, 209–213 (2013).
Barzel, A., Obolski, U., Gogarten, J. P., Kupiec, M. & Hadany, L. Home and away — the evolutionary dynamics of homing endonucleases. BMC Evol. Biol. 11, 324 (2011).
Smillie, C. S. et al. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480, 241–244 (2011). This letter investigates the frequency of HGT in the human microbiome across body sites and across continents.
Rankin, D. J., Rocha, E. P. C. & Brown, S. P. What traits are carried on mobile genetic elements, and why? Hered. (Edinb.). 106, 1–10 (2011). This paper investigates the types of traits that are associated with compound selfish genetic elements and investigates the ecological scenarios that would select for specific types of traits.
Broaders, E., Gahan, C. G. M. & Marchesi, J. R. Mobile genetic elements of the human gastrointestinal tract: potential for spread of antibiotic resistance genes. Gut Microbes 4, 271–280 (2013).
Feschotte, C. & Gilbert, C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet. 13, 283–296 (2012).
Cornelis, G. et al. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc. Natl Acad. Sci. USA 109, E432–E441 (2012).
Schaack, S., Gilbert, C. & Feschotte, C. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol. Evol. 25, 537–546 (2010).
Skippington, E. & Ragan, M. A. Phylogeny rather than ecology or lifestyle biases the construction of Escherichia coli–Shigella genetic exchange communities. Open Biol. 2, 120112 (2012).
Hendrickson, H. & Lawrence, J. G. Selection for chromosome architecture in bacteria. J. Mol. Evol. 62, 615–629 (2006).
Papke, R. T. & Gogarten, J. P. Ecology. How bacterial lineages emerge. Science 336, 45–46 (2012).
Khomyakova, M., Bükmez, Ö., Thomas, L. K., Erb, T. J. & Berg, I. A. A methylaspartate cycle in haloarchaea. Science 331, 334–337 (2011).
Nelson-Sathi, S. et al. Acquisition of 1,000 eubacterial genes physiologically transformed a methanogen at the origin of haloarchaea. Proc. Natl Acad. Sci. USA 109, 20537–20542 (2012).
Nelson-Sathi, S. et al. Origins of major archaeal clades correspond to gene acquisitions from bacteria. Nature 517, 77–80 (2014). This paper suggests that acquisitions of genes from bacteria lead to the evolution of the major clades in archaea.
Guerrero, R., Margulis, L. & Berlanga, M. Symbiogenesis: the holobiont as a unit of evolution. Int. Microbiol. 16, 133–143 (2013).
Ley, R. E., Peterson, D. A. & Gordon, J. I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124, 837–848 (2006).
Hehemann, J.-H. et al. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 464, 908–912 (2010).
Thomas, F. et al. Characterization of the first alginolytic operons in a marine bacterium: from their emergence in marine Flavobacteriia to their independent transfers to marine Proteobacteria and human gut Bacteroides. Environ. Microbiol. 14, 2379–2394 (2012).
Hirt, R. P., Alsmark, C. & Embley, T. M. Lateral gene transfers and the origins of the eukaryote proteome: a view from microbial parasites. Curr. Opin. Microbiol. 23, 155–162 (2015).
McFadden, G. I. Origin and evolution of plastids and photosynthesis in eukaryotes. Cold Spring Harb. Perspect. Biol. 6, a016105 (2014).
Ball, S. G. et al. Metabolic effectors secreted by bacterial pathogens: essential facilitators of plastid endosymbiosis? Plant Cell 25, 7–21 (2013).
Moustafa, A., Reyes-Prieto, A. & Bhattacharya, D. Chlamydiae has contributed at least 55 genes to Plantae with predominantly plastid functions. PLoS ONE 3, e2205 (2008).
Price, D. C. et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science 335, 843–847 (2012).
Cenci, U. et al. Transition from glycogen to starch metabolism in archaeplastida. Trends Plant Sci. 19, 18–28 (2014).
Deschamps, P. Primary endosymbiosis: have cyanobacteria and Chlamydiae ever been roommates? Acta Soc. Bot. Pol. 83, 291–302 (2014).
Ku, C. et al. Endosymbiotic gene transfer from prokaryotic pangenomes: inherited chimerism in eukaryotes. Proc. Natl Acad. Sci. USA http://dx.doi.org/10.1073/pnas.1421385112 (2015).
Domman, D., Horn, M., Embley, T. M. & Williams, T. A. Plastid establishment did not require a chlamydial partner. Nat. Commun. 6, 6421 (2015).
Ball, S. G. et al. Toward an understanding of the function of Chlamydiales in plastid endosymbiosis. Biochim. Biophys. Acta 1847, 495–504 (2015).
Huang, J. & Gogarten, J. P. Concerted gene recruitment in early plant evolution. Genome Biol. 9, R109 (2008).
Suzuki, K. & Miyagishima, S. Y. Eukaryotic and eubacterial contributions to the establishment of plastid proteome estimated by large-scale phylogenetic analyses. Mol. Biol. Evol. 27, 581–590 (2010).
Qiu, H. et al. Assessing the bacterial contribution to the plastid proteome. Trends Plant Sci. 18, 680–687 (2013).
Schonknecht, G. et al. Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339, 1207–1210 (2013).
Blanc, G. et al. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biol. 13, R39 (2012).
Bhattacharya, D. et al. Genome of the red alga Porphyridium purpureum. Nat. Commun. 4, 1941 (2013).
Yue, J., Hu, X. & Huang, J. Origin of plant auxin biosynthesis. Trends Plant Sci. 19, 764–770 (2014).
Yang, Z. et al. Ancient horizontal transfer of transaldolase-like protein gene and its role in plant vascular development. New Phytol. 206, 807–816 (2015).
Hoang, Q. T. et al. An actinoporin plays a key role in water stress in the moss Physcomitrella patens. New Phytol. 184, 502–510 (2009).
Maumus, F., Epert, A., Nogue, F. & Blanc, G. Plant genomes enclose footprints of past infections by giant virus relatives. Nat. Commun. 5, 4268 (2014).
Molbak, L., Molin, S. & Kroer, N. Root growth and exudate production define the frequency of horizontal plasmid transfer in the rhizosphere. FEMS Microbiol. Ecol. 59, 167–176 (2007).
Sun, G., Yang, Z., Ishwar, A. & Huang, J. Algal genes in the closest relatives of animals. Mol. Biol. Evol. 27, 2879–2889 (2010).
Boschetti, C. et al. Biochemical diversification through foreign gene expression in Bdelloid Rotifers. PLoS Genet. 8, e1003035 (2012).
Ricard, G. et al. Horizontal gene transfer from bacteria to rumen ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genomics 7, 22 (2006).
Paganini, J. et al. Contribution of lateral gene transfers to the genome composition and parasitic ability of root-knot nematodes. PLoS ONE 7, e50875 (2012).
Richards, T. A. et al. Horizontal gene transfer facilitated the evolution of plant parasitic mechanisms in the oomycetes. Proc. Natl Acad. Sci. USA 108, 15258–15263 (2011).
Li, Z. W., Shen, Y. H., Xiang, Z. H. & Zhang, Z. Pathogen–origin horizontally transferred genes contribute to the evolution of Lepidopteran insects. BMC Evol. Biol. 11, 356 (2011).
Wybouw, N. et al. A gene horizontally transferred from bacteria protects arthropods from host plant cyanide poisoning. eLife 3, e02365 (2014).
Yoshida, S., Maruyama, S., Nozaki, H. & Shirasu, K. Horizontal gene transfer by the parasitic plant Striga hermonthica. Science 328, 1128 (2010).
Xi, Z. et al. Horizontal transfer of expressed genes in a parasitic flowering plant. BMC Genomics 13, 227 (2012).
Zhang, Y. et al. Evolution of a horizontally acquired legume gene, albumin 1, in the parasitic plant Phelipanche aegyptiaca and related species. BMC Evol. Biol. 13, 48 (2013).
Zhang, D. et al. Root parasitic plant Orobanche aegyptiaca and shoot parasitic plant Cuscuta australis obtained Brassicaceae-specific strictosidine synthase-like genes by horizontal gene transfer. BMC Plant Biol. 14, 19 (2014).
Christin, P.-A. et al. Adaptive evolution of C4 photosynthesis through recurrent lateral gene transfer. Curr. Biol. 22, 445–449 (2012).
Li, F.-W. et al. Horizontal transfer of an adaptive chimeric photoreceptor from bryophytes to ferns. Proc. Natl Acad. Sci. USA 111, 6672–6677 (2014).
Zhang, H.-H., Feschotte, C., Han, M.-J. & Zhang, Z. Recurrent horizontal transfers of Chapaev transposons in diverse invertebrate and vertebrate animals. Genome Biol. Evol. 6, 1375–1386 (2014).
Blanchard, J. L. & Lynch, M. Organellar genes. Trends Genet. 16, 315–320 (2000).
Lynch, M. Mutation accumulation in transfer RNAs: molecular evidence for Muller's ratchet in mitochondrial genomes. Mol. Biol. Evol. 13, 209–220 (1996).
Palmer, J. D. et al. Dynamic evolution of plant mitochondrial genomes: mobile genes and introns and highly variable mutation rates. Proc. Natl Acad. Sci. USA 97, 6960–6966 (2000).
Rice, D. W. et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 342, 1468–1473 (2013). This paper reports the acquisition of several mitochondrial genomes by the mitochondria in Amborella trichopoda a basal flowering plant.
Edwards, A. W. F. Cogwheels of the Mind: The Story of Venn Diagrams (JHU Press, 2004).
Morris, J. J., Lenski, R. E. & Zinser, E. R. The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio 3, e00036-12 (2012). This paper explains how the interplay between cheating and selection for streamlined genomes can give rise to shared genomic resources.
Lobkovsky, A. E., Wolf, Y. I. & Koonin, E. V. Estimation of prokaryotic supergenome size and composition from gene frequency distributions. BMC Genomics 15, S14 (2014).
Lapierre, P. & Gogarten, J. P. Estimating the size of the bacterial pan-genome. Trends Genet. 25, 107–110 (2009).
Puigbò, P., Lobkovsky, A. E., Kristensen, D. M., Wolf, Y. I. & Koonin, E. V. Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Biol. 12, 66 (2014).
Baumdicker, F., Hess, W. R. & Pfaffelhuber, P. The infinitely many genes model for the distributed genome of bacteria. Genome Biol. Evol. 4, 443–456 (2012).
Hooper, L. V. & Gordon, J. I. Commensal host–bacterial relationships in the gut. Science 292, 1115–1118 (2001).
Lederberg, J. & McCray, A. 'Ome sweet 'omics — a genealogical treasury of words. Scientist 15, 8 (2001).
Becker, E. A. et al. Phylogenetically driven sequencing of extremely halophilic archaea reveals strategies for static and dynamic osmo-response. PLoS Genet. 10, e1004784 (2014).
Groussin, M et al. Origins of major archaeal clades do not correspond to gene acquisitions from bacteria. BioRxiv http://dx.doi:10.1101/019851 (2015).
Acknowledgements
Work in the authors' laboratories was supported through grants from the National Science Foundation Grant (DEB 0830024), NASA Exobiology (NNX13AI03G), Binational Science Foundation (BSF 2013061), NSFC Oversea, Hong Kong, Macao collaborative grant (31328003), and the CAS/SAFEA International Partnership Program for Creative Research Teams. The authors would also like to thank K. Swithers for providing insightful comments and discussion pertaining to the body of this text.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information S1 (table)
Summary of Horizontal Gene Transfers (HGTs) discussed in the main text. (PDF 173 kb)
Glossary
- Selfish genetic element
-
A gene or group of genes that enhance their own transmission and reproductive success without making a positive contribution to the host's fitness.
- Microbiome
-
Following a definition ascribed to Joshua Lederberg this term is most often used to denote the collective genome of the indigenous microorganisms of a multicellular or unicellular host. However, the term has also been used by Lederberg and others to signify an ecological community of commensal, symbiotic and pathogenic microorganisms.
- Phylogenetic conflict
-
Differences between the evolutionary history of a species and the evolutionary history of its genes are embodied by discrepancies in branching order between the species and the gene tree.
- Genome streamlining
-
The reduction of genome size through relaxed selection and eventual loss of loci that are superfluous to the niche occupied by the organism.
- Mobilome
-
The aggregate of mobile genetic elements in a genome, population or environment of interest.
- Genome architecture imparting sequences
-
Strand-biased sequence motifs that are enriched towards the termini of replication; thought to direct proteins towards the termini.
- Ecotypes
-
Genetically distinct subsets of organisms within a population or species, usually genetic differences correspond to niche adaptation.
- Holobiont
-
A multicellular or unicellular host and its collective symbionts.
Rights and permissions
About this article
Cite this article
Soucy, S., Huang, J. & Gogarten, J. Horizontal gene transfer: building the web of life. Nat Rev Genet 16, 472–482 (2015). https://doi.org/10.1038/nrg3962
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3962
This article is cited by
-
Host genotype, soil composition, and geo-climatic factors shape the fonio seed microbiome
Microbiome (2024)
-
Metagenomics reveals the temporal dynamics of the rumen resistome and microbiome in goat kids
Microbiome (2024)
-
Horizontal gene transfer is predicted to overcome the diversity limit of competing microbial species
Nature Communications (2024)
-
Horizontal gene transfer in eukaryotes: aligning theory with data
Nature Reviews Genetics (2024)
-
An updated review on how biochar may possess potential in soil ARGs control on aspects of source, fate and elimination
Biochar (2024)