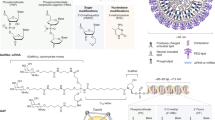
Key Points
-
Synthetic delivery vectors have the potential to address many of the limitations of viral vectors, particularly with respect to safety.
-
For systemic delivery of DNA, both lipid-based vectors and polymer-based vectors have been intensively investigated in experimental animals and in clinical trials.
-
The potential of mRNA for therapeutic protein expression in vivo has been investigated as an alternative to DNA-based gene therapy owing to its unique advantages. Recent advances in chemical modifications of mRNA reduce stimulation of the immune system and improve stability when it is delivered in vivo.
-
Small interfering RNA (siRNA) has great therapeutic potential, as it can silence nearly any targeted gene after introduction into cells. Lipid- and polymer-based siRNA nanoparticles and conjugate systems enable successful delivery of chemically modified siRNAs in humans.
-
Levels of microRNA (miRNA) can be restored through the introduction of synthetic miRNAs or mimics as miRNA replacement therapy. One miRNA mimic is currently undergoing evaluation in a Phase I clinical trial.
-
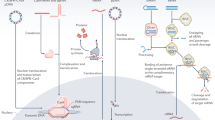
Delivery of genome editing systems — including zinc-finger proteins, transcription activator-like effectors and CRISPR–Cas (clustered regularly interspaced short palindromic repeat–CRISPR-associated) systems — facilitates gene editing at desired sites in the genome. Recent proof-of-concept studies in model organisms have shown that this approach may be used to cure genetic diseases, which is in contrast to the temporary expression or random insertion of a DNA fragment in conventional gene therapy.
Abstract
Gene-based therapy is the intentional modulation of gene expression in specific cells to treat pathological conditions. This modulation is accomplished by introducing exogenous nucleic acids such as DNA, mRNA, small interfering RNA (siRNA), microRNA (miRNA) or antisense oligonucleotides. Given the large size and the negative charge of these macromolecules, their delivery is typically mediated by carriers or vectors. In this Review, we introduce the biological barriers to gene delivery in vivo and discuss recent advances in material sciences, nanotechnology and nucleic acid chemistry that have yielded promising non-viral delivery systems, some of which are currently undergoing testing in clinical trials. The diversity of these systems highlights the recent progress of gene-based therapy using non-viral approaches.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ginn, S. L., Alexander, I. E., Edelstein, M. L., Abedi, M. R. & Wixon, J. Gene therapy clinical trials worldwide to 2012 — an update. J. Gene Med. 15, 65–77 (2013).
Kay, M. A. State-of-the-art gene-based therapies: the road ahead. Nature Rev. Genet. 12, 316–328 (2011).
Mingozzi, F. & High, K. A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nature Rev. Genet. 12, 341–355 (2011).
Baum, C., Kustikova, O., Modlich, U., Li, Z. & Fehse, B. Mutagenesis and oncogenesis by chromosomal insertion of gene transfer vectors. Hum. Gene Ther. 17, 253–263 (2006).
Bessis, N., GarciaCozar, F. J. & Boissier, M. C. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 11 (Suppl. 1), S10–S17 (2004).
Waehler, R., Russell, S. J. & Curiel, D. T. Engineering targeted viral vectors for gene therapy. Nature Rev. Genet. 8, 573–587 (2007).
Thomas, C. E., Ehrhardt, A. & Kay, M. A. Progress and problems with the use of viral vectors for gene therapy. Nature Rev. Genet. 4, 346–358 (2003).
Bouard, D., Alazard-Dany, D. & Cosset, F. L. Viral vectors: from virology to transgene expression. Br. J. Pharmacol. 157, 153–165 (2009).
Pack, D. W., Hoffman, A. S., Pun, S. & Stayton, P. S. Design and development of polymers for gene delivery. Nature Rev. Drug Discov. 4, 581–593 (2005).
Mintzer, M. A. & Simanek, E. E. Nonviral vectors for gene delivery. Chem. Rev. 109, 259–302 (2009).
Putnam, D. Polymers for gene delivery across length scales. Nature Mater. 5, 439–451 (2006).
Davis, M. E. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol. Pharmaceut. 6, 659–668 (2009).
Gonzalez, H., Hwang, S. J. & Davis, M. E. New class of polymers for the delivery of macromolecular therapeutics. Bioconjug. Chem. 10, 1068–1074 (1999).
Semple, S. C. et al. Rational design of cationic lipids for siRNA delivery. Nature Biotech. 28, 172–176 (2010). This study carries out a rational design of a siRNA delivery vector and achieved significant improvement in delivery efficiency.
Love, K. T. et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl Acad. Sci. USA 107, 1864–1869 (2010).
Monopoli, M. P., Aberg, C., Salvati, A. & Dawson, K. A. Biomolecular coronas provide the biological identity of nanosized materials. Nature Nanotechnol. 7, 779–786 (2012).
Lee, H. et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nature Nanotechnol. 7, 389–393 (2012).
Kariko, K., Buckstein, M., Ni, H. & Weissman, D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005).
Anderson, B. R. et al. Nucleoside modifications in RNA limit activation of 2′-5′-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 39, 9329–9338 (2011).
Kariko, K., Muramatsu, H., Keller, J. M. & Weissman, D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 20, 948–953 (2012).
Deleavey, G. F., Watts, J. K. & Damha, M. J. Chemical modification of siRNA. Curr. Protoc. Nucleic Acid Chem. 39, 16.3.1–16.3.22 (2009).
Mehier-Humbert, S. & Guy, R. H. Physical methods for gene transfer: improving the kinetics of gene delivery into cells. Adv. Drug Deliv. Rev. 57, 733–753 (2005).
Wells, D. J. Gene therapy progress and prospects: electroporation and other physical methods. Gene Ther. 11, 1363–1369 (2004).
Newman, C. M. & Bettinger, T. Gene therapy progress and prospects: ultrasound for gene transfer. Gene Ther. 14, 465–475 (2007).
Plank, C. et al. The magnetofection method: using magnetic force to enhance gene delivery. Biol. Chem. 384, 737–747 (2003).
Zhang, G., Budker, V. G., Ludtke, J. J. & Wolff, J. A. Naked DNA gene transfer in mammalian cells. Methods Mol. Biol. 245, 251–264 (2004).
Li, W. & Szoka, F. C. Jr. Lipid-based nanoparticles for nucleic acid delivery. Pharm. Res. 24, 438–449 (2007).
Thomas, M. & Klibanov, A. M. Non-viral gene therapy: polycation-mediated DNA delivery. Appl. Microbiol. Biotechnol. 62, 27–34 (2003).
Lee, C. C., MacKay, J. A., Frechet, J. M. & Szoka, F. C. Designing dendrimers for biological applications. Nature Biotech. 23, 1517–1526 (2005).
Discher, D. E. & Ahmed, F. Polymersomes. Annu. Rev. Biomed. Engineer. 8, 323–341 (2006).
Martin, M. E. & Rice, K. G. Peptide-guided gene delivery. AAPS J. 9, E18–29 (2007).
Sokolova, V. & Epple, M. Inorganic nanoparticles as carriers of nucleic acids into cells. Angew. Chem. Int. Ed. Engl. 47, 1382–1395 (2008).
Kawabata, K., Takakura, Y. & Hashida, M. The fate of plasmid DNA after intravenous injection in mice: involvement of scavenger receptors in its hepatic uptake. Pharm. Res. 12, 825–830 (1995).
McManus, J. J., Radler, J. O. & Dawson, K. A. Observation of a rectangular columnar phase in a DNA–calcium–zwitterionic lipid complex. J. Am. Chem. Soc. 126, 15966–15967 (2004).
McManus, J. J., Rädler, J. O. & Dawson, K. A. Does calcium turn a zwitterionic lipid cationic? J. Phys. Chem. B 107, 9869–9875 (2003).
Koltover, I., Salditt, T., Radler, J. O. & Safinya, C. R. An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science 281, 78–81 (1998). This paper elucidates structural considerations involved in the endosomal escape of DNA mediated by liposomal nanoparticles.
Wiethoff, C. M. & Middaugh, C. R. Barriers to nonviral gene delivery. J. Pharm. Sci. 92, 203–217 (2003).
Morille, M., Passirani, C., Vonarbourg, A., Clavreul, A. & Benoit, J. P. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials 29, 3477–3496 (2008).
Capecchi, M. R. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell 22, 479–488 (1980).
Miller, A. M. & Dean, D. A. Tissue-specific and transcription factor-mediated nuclear entry of DNA. Adv. Drug Deliv. Rev. 61, 603–613 (2009).
Varga, C. M. et al. Quantitative comparison of polyethylenimine formulations and adenoviral vectors in terms of intracellular gene delivery processes. Gene Ther. 12, 1023–1032 (2005).
Dinh, A. T., Pangarkar, C., Theofanous, T. & Mitragotri, S. Understanding intracellular transport processes pertinent to synthetic gene delivery via stochastic simulations and sensitivity analyses. Biophys. J. 92, 831–846 (2007).
Wasungu, L. & Hoekstra, D. Cationic lipids, lipoplexes and intracellular delivery of genes. J. Control. Release 116, 255–264 (2006).
Godbey, W. T., Wu, K. K. & Mikos, A. G. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc. Natl Acad. Sci. USA 96, 5177–5181 (1999).
Breunig, M. et al. Gene delivery with low molecular weight linear polyethylenimines. J. Gene Med. 7, 1287–1298 (2005).
Schaffer, D. V., Fidelman, N. A., Dan, N. & Lauffenburger, D. A. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol. Bioeng. 67, 598–606 (2000).
Cohen, R. N., van der Aa, M. A., Macaraeg, N., Lee, A. P. & Szoka, F. C. Jr. Quantification of plasmid DNA copies in the nucleus after lipoplex and polyplex transfection. J. Controlled Release 135, 166–174 (2009).
Gill, D. R., Pringle, I. A. & Hyde, S. C. Progress and prospects: the design and production of plasmid vectors. Gene Ther. 16, 165–171 (2009).
Gill, D. R. et al. Increased persistence of lung gene expression using plasmids containing the ubiquitin C or elongation factor 1α promoter. Gene Ther. 8, 1539–1546 (2001).
Wooddell, C. I., Reppen, T., Wolff, J. A. & Herweijer, H. Sustained liver-specific transgene expression from the albumin promoter in mice following hydrodynamic plasmid DNA delivery. J. Gene Med. 10, 551–563 (2008).
Miao, C. H. et al. Inclusion of the hepatic locus control region, an intron, and untranslated region increases and stabilizes hepatic factor IX gene expression in vivo but not in vitro. Mol. Ther. 1, 522–532 (2000).
Argyros, O. et al. Persistent episomal transgene expression in liver following delivery of a scaffold/matrix attachment region containing non-viral vector. Gene Ther. 15, 1593–1605 (2008).
Kreiss, P. et al. Plasmid DNA size does not affect the physicochemical properties of lipoplexes but modulates gene transfer efficiency. Nucleic Acids Res. 27, 3792–3798 (1999).
Darquet, A. M. et al. Minicircle: an improved DNA molecule for in vitro and in vivo gene transfer. Gene Ther. 6, 209–218 (1999).
Kay, M. A., He, C. Y. & Chen, Z. Y. A robust system for production of minicircle DNA vectors. Nature Biotech. 28, 1287–1289 (2010).
Ehrhardt, A., Xu, H., Huang, Z., Engler, J. A. & Kay, M. A. A direct comparison of two nonviral gene therapy vectors for somatic integration: in vivo evaluation of the bacteriophage integrase phiC31 and the Sleeping Beauty transposase. Mol. Ther. 11, 695–706 (2005).
Wu, S. C. et al. PiggyBac is a flexible and highly active transposon as compared to Sleeping Beauty, Tol2, and Mos1 in mammalian cells. Proc. Natl Acad. Sci. USA 103, 15008–15013 (2006).
Aronovich, E. L., McIvor, R. S. & Hackett, P. B. The Sleeping Beauty transposon system: a non-viral vector for gene therapy. Hum. Mol. Genet. 20, R14–R20 (2011).
Fraley, R., Subramani, S., Berg, P. & Papahadjopoulos, D. Introduction of liposome-encapsulated SV40 DNA into cells. J. Biol. Chem. 255, 10431–10435 (1980).
Felgner, P. L. et al. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl Acad. Sci. USA 84, 7413–7417 (1987). References 59 and 60 are the earliest studies to show lipid-mediated DNA delivery in vitro.
Whitehead, K. A., Langer, R. & Anderson, D. G. Knocking down barriers: advances in siRNA delivery. Nature Rev. Drug Discov. 8, 129–138 (2009).
Lonez, C., Vandenbranden, M. & Ruysschaert, J. M. Cationic liposomal lipids: from gene carriers to cell signaling. Prog. Lipid Res. 47, 340–347 (2008).
Hersey, P. & Gallagher, S. Intralesional immunotherapy for melanoma. J. Surg. Oncol. 109, 320–326 (2014).
Olins, D. E., Olins, A. L. & Von Hippel, P. H. Model nucleoprotein complexes: studies on the interaction of cationic homopolypeptides with DNA. J. Mol. Biol. 24, 157–176 (1967).
Laemmli, U. K. Characterization of DNA condensates induced by poly(ethylene oxide) and polylysine. Proc. Natl Acad. Sci. USA 72, 4288–4292 (1975).
Wu, G. Y. & Wu, C. H. Receptor-mediated in vitro gene transformation by a soluble DNA carrier system. J. Biol. Chem. 262, 4429–4432 (1987).
Wu, G. Y. & Wu, C. H. Receptor-mediated gene delivery and expression in vivo. J. Biol. Chem. 263, 14621–14624 (1988). References 66 and 67 are among the first to investigate the possibility of targeted non-viral nucleic acid delivery in vitro and in vivo.
Choi, Y. H. et al. Polyethylene glycol-grafted poly-L-lysine as polymeric gene carrier. J. Control. Release 54, 39–48 (1998).
Kim, S. W. Polylysine copolymers for gene delivery. Cold Spring Harb.Protoc. 2012, 433–438 (2012).
Alexis, F., Pridgen, E., Molnar, L. K. & Farokhzad, O. C. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol. Pharm. 5, 505–515 (2008).
Bazile, D. et al. Stealth Me. PEG–PLA nanoparticles avoid uptake by the mononuclear phagocytes system. J. Pharm. Sci. 84, 493–498 (1995).
Konstan, M. W. et al. Compacted DNA nanoparticles administered to the nasal mucosa of cystic fibrosis subjects are safe and demonstrate partial to complete cystic fibrosis transmembrane regulator reconstitution. Hum. Gene Ther. 15, 1255–1269 (2004).
Lungwitz, U., Breunig, M., Blunk, T. & Gopferich, A. Polyethylenimine-based non-viral gene delivery systems. Eur. J. Pharm. Biopharm. 60, 247–266 (2005).
Boussif, O. et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl Acad. Sci. USA 92, 7297–7301 (1995). This is the first paper to show that PEI can facilitate DNA transfection in vitro and in vivo.
Godbey, W. T., Wu, K. K. & Mikos, A. G. Size matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J. Biomed. Mater. Res. 45, 268–275 (1999).
Wightman, L. et al. Different behavior of branched and linear polyethylenimine for gene delivery in vitro and in vivo. J. Gene Med. 3, 362–372 (2001).
Goula, D. et al. Polyethylenimine-based intravenous delivery of transgenes to mouse lung. Gene Ther. 5, 1291–1295 (1998).
Kircheis, R., Wightman, L. & Wagner, E. Design and gene delivery activity of modified polyethylenimines. Adv. Drug Deliv. Rev. 53, 341–358 (2001).
Coll, J. L. et al. In vivo delivery to tumors of DNA complexed with linear polyethylenimine. Hum. Gene Ther. 10, 1659–1666 (1999).
Lv, H., Zhang, S., Wang, B., Cui, S. & Yan, J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release 114, 100–109 (2006).
Nguyen, H. K. et al. Evaluation of polyether-polyethyleneimine graft copolymers as gene transfer agents. Gene Ther. 7, 126–138 (2000).
Petersen, H. et al. Polyethylenimine-graft-poly(ethylene glycol) copolymers: influence of copolymer block structure on DNA complexation and biological activities as gene delivery system. Bioconjug. Chem. 13, 845–854 (2002).
Breunig, M., Lungwitz, U., Liebl, R. & Goepferich, A. Breaking up the correlation between efficacy and toxicity for nonviral gene delivery. Proc. Natl Acad. Sci. USA 104, 14454–14459 (2007).
Thomas, M. & Klibanov, A. M. Enhancing polyethylenimine's delivery of plasmid DNA into mammalian cells. Proc. Natl Acad. Sci. USA 99, 14640–14645 (2002).
Fortune, J. A., Novobrantseva, T. I. & Klibanov, A. M. Highly effective gene transfection in vivo by alkylated polyethylenimine. J. Drug Deliv. 2011, 204058 (2011).
Zangi, L. et al. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nature Biotech. 31, 898–907 (2013).
Kormann, M. S. et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nature Biotech. 29, 154–157 (2011). References 86 and 87 show the therapeutic potential of delivery of modified mRNA in vivo.
Su, X., Fricke, J., Kavanagh, D. G. & Irvine, D. J. In vitro and in vivo mRNA delivery using lipid-enveloped pH-responsive polymer nanoparticles. Mol. Pharm. 8, 774–787 (2011).
Phua, K. K., Leong, K. W. & Nair, S. K. Transfection efficiency and transgene expression kinetics of mRNA delivered in naked and nanoparticle format. J. Control. Release 166, 227–233 (2013).
Kanasty, R. L., Whitehead, K. A., Vegas, A. J. & Anderson, D. G. Action and reaction: the biological response to siRNA and its delivery vehicles. Mol. Ther. 20, 513–524 (2012).
Layzer, J. M. et al. In vivo activity of nuclease-resistant siRNAs. RNA 10, 766–771 (2004).
Jackson, A. L. & Linsley, P. S. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nature Rev. Drug Discov. 9, 57–67 (2010).
Nguyen, D. N. et al. Lipid-derived nanoparticles for immunostimulatory RNA adjuvant delivery. Proc. Natl Acad. Sci. USA 109, E797–E803 (2012).
Whitehead, K. A., Dahlman, J. E., Langer, R. S. & Anderson, D. G. Silencing or stimulation? siRNA delivery and the immune system. Annu. Rev. Chem. Biomol. Eng. 2, 77–96 (2011).
Wang, A. Z., Langer, R. & Farokhzad, O. C. Nanoparticle delivery of cancer drugs. Annu. Rev. Med. 63, 185–198 (2012).
Wolfrum, C. et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nature Biotech. 25, 1149–1157 (2007).
Akinc, A. et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 18, 1357–1364 (2010).
Malek, A. et al. In vivo pharmacokinetics, tissue distribution and underlying mechanisms of various PEI(-PEG)/siRNA complexes. Toxicol. Appl. Pharmacol. 236, 97–108 (2009).
Huang, Y. et al. Elimination pathways of systemically delivered siRNA. Mol. Ther. 19, 381–385 (2011).
Rozema, D. B. et al. Dynamic PolyConjugates for targeted in vivo delivery of siRNA to hepatocytes. Proc. Natl Acad. Sci. USA 104, 12982–12987 (2007).
Kanasty, R., Dorkin, J. R., Vegas, A. & Anderson, D. Delivery materials for siRNA therapeutics. Nature Mater. 12, 967–977 (2013).
Zuckerman, J. E., Choi, C. H., Han, H. & Davis, M. E. Polycation–siRNA nanoparticles can disassemble at the kidney glomerular basement membrane. Proc. Natl Acad. Sci. USA 109, 3137–3142 (2012).
Naeye, B. et al. In vivo disassembly of IV administered siRNA matrix nanoparticles at the renal filtration barrier. Biomaterials 34, 2350–2358 (2013).
Aird, W. C. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circul. Res. 100, 158–173 (2007).
Wisse, E., Jacobs, F., Topal, B., Frederik, P. & De Geest, B. The size of endothelial fenestrae in human liver sinusoids: implications for hepatocyte-directed gene transfer. Gene Ther. 15, 1193–1199 (2008).
Maeda, H. Tumor-selective delivery of macromolecular drugs via the EPR effect: background and future prospects. Bioconjug. Chem. 21, 797–802 (2010).
Yu, B., Zhao, X., Lee, L. J. & Lee, R. J. Targeted delivery systems for oligonucleotide therapeutics. AAPS J. 11, 195–203 (2009).
Salvati, A. et al. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nature Nanotechnol. 8, 137–143 (2013).
Sahay, G. et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nature Biotech. 31, 653–658 (2013). This study elucidates the mechanism of cellular uptake of LNPs and the importance of exocytosis in limiting delivery efficiency.
Schwarz, D. S. et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208 (2003).
Geisbert, T. W. et al. Postexposure protection of non-human primates against a lethal Ebola virus challenge with RNA interference: a proof-of-concept study. Lancet 375, 1896–1905 (2010).
Zimmermann, T. S. et al. RNAi-mediated gene silencing in non-human primates. Nature 441, 111–114 (2006).
Wheeler, J. J. et al. Stabilized plasmid–lipid particles: construction and characterization. Gene Ther. 6, 271–281 (1999).
Morrissey, D. V. et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nature Biotech. 23, 1002–1007 (2005).
Jayaraman, M. et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. Engl. 51, 8529–8533 (2012).
Akinc, A. et al. Development of lipidoid–siRNA formulations for systemic delivery to the liver. Mol. Ther. 17, 872–879 (2009).
Alabi, C. A. et al. Multiparametric approach for the evaluation of lipid nanoparticles for siRNA delivery. Proc. Natl Acad. Sci. USA 110, 12881–12886 (2013).
Burnett, J. C., Rossi, J. J. & Tiemann, K. Current progress of siRNA/shRNA therapeutics in clinical trials. Biotechnol. J. 6, 1130–1146 (2011).
Fitzgerald, K. et al. Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. Lancet 383, 60–68 (2014).
Santel, A. et al. A novel siRNA–lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 13, 1222–1234 (2006).
Santel, A. et al. RNA interference in the mouse vascular endothelium by systemic administration of siRNA–lipoplexes for cancer therapy. Gene Ther. 13, 1360–1370 (2006).
Aleku, M. et al. Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res. 68, 9788–9798 (2008).
Landen, C. N. Jr. et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 65, 6910–6918 (2005).
Davis, M. E. et al. Self-assembling nucleic acid delivery vehicles via linear, water-soluble, cyclodextrin-containing polymers. Curr. Med. Chem. 11, 179–197 (2004).
Hu-Lieskovan, S., Heidel, J. D., Bartlett, D. W., Davis, M. E. & Triche, T. J. Sequence-specific knockdown of EWS–FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing's sarcoma. Cancer Res. 65, 8984–8992 (2005).
Bartlett, D. W. & Davis, M. E. Impact of tumor-specific targeting and dosing schedule on tumor growth inhibition after intravenous administration of siRNA-containing nanoparticles. Biotechnol. Bioeng. 99, 975–985 (2008).
Heidel, J. D. et al. Administration in non-human primates of escalating intravenous doses of targeted nanoparticles containing ribonucleotide reductase subunit M2 siRNA. Proc. Natl Acad. Sci. USA 104, 5715–5721 (2007).
Davis, M. E. et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature 464, 1067–1070 (2010). This paper provides the first evidence that systemically administered siRNA can induce RNAi in humans.
Wong, S. C. et al. Co-injection of a targeted, reversibly masked endosomolytic polymer dramatically improves the efficacy of cholesterol-conjugated small interfering RNAs in vivo. Nucleic Acid. Ther. 22, 380–390 (2012).
Rozema, D. B., Ekena, K., Lewis, D. L., Loomis, A. G. & Wolff, J. A. Endosomolysis by masking of a membrane-active agent (EMMA) for cytoplasmic release of macromolecules. Bioconjug.Chem. 14, 51–57 (2003).
Wooddell, C. I. et al. Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Mol. Ther. 21, 973–985 (2013).
Yasuda, M. et al. RNAi-mediated silencing of hepatic Alas1 effectively prevents and treats the induced acute attacks in acute intermittent porphyria mice. Proc. Natl Acad. Sci. USA http://dx.doi.org/10.1073/pnas.1406228111 (2014).
Chen, K. & Rajewsky, N. The evolution of gene regulation by transcription factors and microRNAs. Nature Rev. Genet. 8, 93–103 (2007).
Pritchard, C. C., Cheng, H. H. & Tewari, M. MicroRNA profiling: approaches and considerations. Nature Rev. Genet. 13, 358–369 (2012).
Hydbring, P., Badalian-Very, G. Clinical applications of microRNAs F1000Res. 2, 136 (2013).
Nana-Sinkam, S. P. & Croce, C. M. Clinical applications for microRNAs in cancer. Clin. Pharmacol. Ther. 93, 98–104 (2013).
He, L. et al. A microRNA component of the p53 tumour suppressor network. Nature 447, 1130–1134 (2007).
Trang, P. et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 19, 1116–1122 (2011).
Bader, A. G. miR-34 — a microRNA replacement therapy is headed to the clinic. Front. Genet. 3, 120 (2012).
Gaj, T., Gersbach, C. A. & Barbas, C. F. 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405 (2013).
Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S. & Gregory, P. D. Genome editing with engineered zinc finger nucleases. Nature Rev. Genet. 11, 636–646 (2010).
Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009).
Moscou, M. J. & Bogdanove, A. J. A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501 (2009).
Miller, J. C. et al. A TALE nuclease architecture for efficient genome editing. Nature Biotech. 29, 143–148 (2011).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Wang, H. et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013).
Ran, F. A. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154, 1380–1389 (2013).
Li, H. et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature 475, 217–221 (2011). This study is the first to show that genome editing tools (ZFN) can modify gene in vivo and rescue disease phenotype.
Yin, H. et al. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nature Biotech. 32, 551–553 (2014). This paper is the first to show that the Cas9 system can correct a disease mutation and rescue disease phenotype in adult animals.
Hopkins, A. L. & Groom, C. R. The druggable genome. Nature Rev. Drug Discov. 1, 727–730 (2002).
Heyes, J., Palmer, L., Bremner, K. & MacLachlan, I. Cationic lipid saturation influences intracellular delivery of encapsulated nucleic acids. J. Control. Release 107, 276–287 (2005).
Ambegia, E. et al. Stabilized plasmid-lipid particles containing PEG–diacylglycerols exhibit extended circulation lifetimes and tumor selective gene expression. Biochim. Biophys. Acta 1669, 155–163 (2005).
Mishra, S., Heidel, J. D., Webster, P. & Davis, M. E. Imidazole groups on a linear, cyclodextrin-containing polycation produce enhanced gene delivery via multiple processes. J. Control. Release 116, 179–191 (2006).
Bellocq, N. C., Pun, S. H., Jensen, G. S. & Davis, M. E. Transferrin-containing, cyclodextrin polymer-based particles for tumor-targeted gene delivery. Bioconjug. Chem. 14, 1122–1132 (2003).
Mishra, S., Webster, P. & Davis, M. E. PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles. Eur. J. Cell Biol. 83, 97–111 (2004).
Ramalingam, S., Annaluru, N. & Chandrasegaran, S. A CRISPR way to engineer the human genome. Genome Biol. 14, 107 (2013).
Acknowledgements
The authors thank R. L. Bogorad, E. A. Appel, O. F. Khan, O. Veiseh, Y. Dong and G. Sahay for discussion. H.Y. is supported by the US National Institutes of Health (NIH) Centers for Cancer Nanotechnology Excellence and the Harvard–MIT (Massachusetts Institute of Technology) Center of Cancer Nanotechnology Excellence (5-U54-CA151884-04). R.K. is supported by the US National Science Foundation Graduate Research Fellowship (grant 1122374). A.A.E. is supported by the US National Heart, Lung, and Blood Institute, NIH, as a Program of Excellence in Nanotechnology (PEN) Award (contract HHSN268201000045C). This work was supported partly by the Koch Institute Support (core) Grant P30-CA14051 from the US National Cancer Institute. The authors acknowledge the service to the MIT community of the late S. Collier.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
D.G.A. is a cofounder of CRISPR Therapeutics. D.G.A. has a research grant with Alnylam Pharmaceuticals and a research grant with Shire. J.R.D. is a shareholder of Alnylam Pharmaceuticals.
Glossary
- Retroviruses
-
Single-stranded RNA viruses that use reverse transcriptase to transcribe their RNA into DNA, which can be integrated into the host genome. The viral DNA can be transcribed, translated and packed into new viruses. Replication-defective retroviruses are commonly used for gene therapy purposes. Most of these retroviruses are only active in dividing cells.
- Lentiviruses
-
A subclass of retroviruses that are active in non-dividing cells. The replication-defective viruses are used both as a research tool to introduce a gene in vitro or in vivo and for gene therapy. They infect cells with high efficiency and introduce stable expression.
- Adenoviruses
-
DNA viruses that do not integrate into the host genome and that usually do not replicate during cell division. Many of them trigger fast immune responses. For gene therapy purposes, they are usually applied in conditions in which temporary expression of proteins is required.
- Adeno-associated viruses
-
(AAVs). DNA viruses that have very low but measurable genome integration rates and that are used as vectors for gene therapy. They are able to infect both dividing and non-dividing cells with high efficiency and long persistence. Although they have small packing capability, they are preferred for gene therapy owing to their low immunogenicity and low cytotoxicity.
- Zinc-finger proteins
-
(ZFPs). DNA-binding proteins that consist of tandem arrays of zinc-fingers, which are protein structure motifs that contain one or more zinc ions. Engineered zinc-fingers have been shown to recognize three specific base pairs of DNA sequences and can be assembled in tandem to recognize specific nucleic acid sequences. The process of engineering is difficult and requires expertise.
- Transcription activator-like effectors
-
(TALEs). Proteins that were first discovered in Xanthomonas spp. bacteria and that bind to promoter sequences in host plants to facilitate infections. They contain a repeat domain of 34 amino acids. Two critical amino acids in each repeat allow targeting of specific DNA bases. TALEs can be engineered in a time-consuming process by assembling repeat domains to recognize specific DNA sequences.
- CRISPR–Cas
-
(Clustered regularly interspaced short palindromic repeat–CRISPR-associated). Defense systems against foreign DNA in bacteria and archaea, in which a short CRISPR RNA (crRNA) is used to guide the Cas nuclease to a specific target DNA sequence. These systems have been optimized to function in mammalian cells with high efficiency. The engineering process to target various DNA sequences is straightforward, and the cost is low.
- Liposomes
-
Vesicles of various sizes with a lipid bilayer that can encapsulate small molecules or large molecules such as small interfering RNA or DNA. By fusing to the cell membrane, they facilitate delivery of the liposome contents in vitro and in vivo.
- Polymersomes
-
Vesicles produced using co-polymers, which are made of two or more monomers, to allow both hydrophilic and lipophilic ability. They can encapsulate small molecules, proteins and DNA to form particles of various sizes, and are used for drug delivery systems in vitro and in vivo.
- Cell-penetrating peptides
-
Small peptides that can translocate across plasma membranes. By covalent or non-covalent binding of these peptides to small molecules, proteins, DNA, small interfering RNA or even nanoparticles, they could facilitate intracellular delivery of various types of molecules.
- Scaffold/matrix attachment regions
-
(S/MARs). Anchor points of the genomic DNA for the chromatin scaffold. They are found at introns or borders of transcription units, where they have an important role in separating these units and regulating gene expression.
- PhiC31
-
A sequence-specific recombinase that mediates recombination between 2 specific 34-bp sequences, which allows insertion of DNA into another DNA molecule or into the genome at specific sites.
- PiggyBac
-
A transposon system composed of a transposon and a transposase, which recognizes transposon-specific sequences on both ends of a vector and integrates the content into different DNA molecules or genome.
- Sleeping Beauty
-
A transposon system reconstructed from DNA copies of salmon. The transposase was engineered to facilitate robust and stable gene transfer.
- RNAiMAX
-
A commercial reagent that delivers small interfering RNAs into various types of cells in vitro with high efficiency. As a cationic lipid formulation, it can also be used to deliver microRNA antagonists and mimics, as well as mRNAs.
- Glomerular filtration barrier
-
A blood filtration interface in the kidney that allows free passage of small ions such as sodium and potassium but that retains large proteins. The cutoff to pass this barrier is ~70 kDa, and naked small interfering RNAs (~13 kDa) can thus be filtered through the kidney.
- FokI
-
A restriction endonuclease with a DNA-binding domain at the amino terminus and a nonspecific DNA cleavage domain at the carboxyl terminus. Dimerization of two FokI is required to cleave DNA. When it is fused to zinc-finger proteins (ZFPs) or to transcription activator-like effectors (TALEs), FokI will function when a pair of ZFPs or TALEs binds to DNA within short distance. This strategy allows increased specificity of sequence recognition.
Rights and permissions
About this article
Cite this article
Yin, H., Kanasty, R., Eltoukhy, A. et al. Non-viral vectors for gene-based therapy. Nat Rev Genet 15, 541–555 (2014). https://doi.org/10.1038/nrg3763
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3763
This article is cited by
-
Recent progress of non-linear topological structure polymers: synthesis, and gene delivery
Journal of Nanobiotechnology (2024)
-
Harnessing cell reprogramming for cardiac biological pacing
Journal of Biomedical Science (2023)
-
Research progress of engineered mesenchymal stem cells and their derived exosomes and their application in autoimmune/inflammatory diseases
Stem Cell Research & Therapy (2023)
-
Dual peptides-modified cationic liposomes for enhanced Lung cancer gene therapy by a gap junction regulating strategy
Journal of Nanobiotechnology (2023)
-
Therapeutic effects of engineered exosome-based miR-25 and miR-181a treatment in spinocerebellar ataxia type 3 mice by silencing ATXN3
Molecular Medicine (2023)