Key Points
-
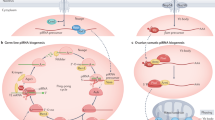
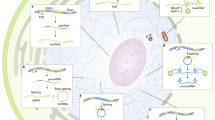
Like nearly all biological mechanisms, small RNA-directed pathways are both elegantly simple — small RNA guides use sequence complementarity to identify their targets — and shockingly complex, with myriad proteins required to excise small RNA guides from much longer precursors and still more required to carry out small RNA-directed functions.
-
Despite this complexity, the defining features of small silencing RNAs are their short length (∼20–30 nucleotides) and their association with members of the Argonaute family of proteins.
-
Small interfering RNAs are typically 21 nucleotides and they are derived from dsRNA, the nearly universal eukaryotic signal for 'foreignness'.
-
Small interfering RNAs can be both exogenous and endogenous in origin in plants and animals, and provide an epigenetic component of resistance against biotic and abiotic stress.
-
MicroRNAs 'tune' and regulate development and many other biological processes in plants and animals. MicroRNAs are encoded in the genome and typically repress their mRNA targets by partial base-pairing, and hence have the potential to regulate many distinct mRNA targets.
-
Piwi-interacting RNAs participate in a feed-forward amplification loop that monitors and silences transposon expression in the germ line.
-
Small RNA pathways, although distinct, share protein components and repress and enhance each other. Such 'cross talk' between small RNA pathways is poorly understood.
Abstract
Since the discovery in 1993 of the first small silencing RNA, a dizzying number of small RNA classes have been identified, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs). These classes differ in their biogenesis, their modes of target regulation and in the biological pathways they regulate. There is a growing realization that, despite their differences, these distinct small RNA pathways are interconnected, and that small RNA pathways compete and collaborate as they regulate genes and protect the genome from external and internal threats.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Fire, A. et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 (1998). The landmark discovery of dsRNA as the trigger for RNAi.
Voinnet, O. Non-cell autonomous RNA silencing. FEBS Lett. 579, 5858–5871 (2005).
Grishok, A., Tabara, H. & Mello, C. C. Genetic requirements for inheritance of RNAi in C. elegans. Science 287, 2494–2497 (2000).
Hamilton, A. J. & Baulcombe, D. C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950–952 (1999). This paper identified small RNAs — now known as siRNAs — as the guides for post-transcriptional gene silencing in plants.
Zamore, P. D., Tuschl, T., Sharp, P. A. & Bartel, D. P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33 (2000). This paper and reference 6 showed that siRNAs are the specificity determinants for endonucleolytic cleavage of RNA targets by RISC.
Hammond, S. M., Bernstein, E., Beach, D. & Hannon, G. J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296 (2000).
Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001). In this work, Dicer was shown to be the dsRNA-specific ribonuclease that converts long dsRNA into siRNAs.
Elbashir, S. M., Lendeckel, W. & Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200 (2001). This paper and reference 9 deduced the structure of siRNAs and discovered that they direct cleavage at the phosphodiester bond in the target that lies across from bases 10 and 11 in the small RNA guide, now a general principle for Argonaute-mediated, small RNA-guided silencing.
Elbashir, S. M., Martinez, J., Patkaniowska, A., Lendeckel, W. & Tuschl, T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20, 6877–6888 (2001).
Liu, J. et al. A role for the P-body component GW182 in microRNA function. Nature Cell Biol. 7, 1161–1166 (2005).
Hutvágner, G. et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838 (2001). This study, together with references 12 and 13, linked the RNAi and miRNA pathways, establishing that they share proteins, such as Dicer, and mechanisms of biogenesis and function.
Grishok, A. et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34 (2001).
Ketting, R. F. et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659 (2001).
Knight, S. W. & Bass, B. L. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293, 2269–2271 (2001).
Lee, Y. S. et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69–81 (2004).
Obbard, D. J., Jiggins, F. M., Halligan, D. L. & Little, T. J. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 16, 580–585 (2006).
Tabara, H., Yigit, E., Siomi, H. & Mello, C. C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DexH-Box helicase to direct RNAi in C. elegans. Cell 109, 861–871 (2002).
Shaham, S. Worming into the cell: viral reproduction in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 103, 3955–3956 (2006).
Williams, B. R. PKR; a sentinel kinase for cellular stress. Oncogene 18, 6112–6120 (1999).
Kunzi, M. S. & Pitha, P. M. Interferon research: a brief history. Methods Mol. Med. 116, 25–35 (2005).
Vilcek, J. Fifty years of interferon research: aiming at a moving target. Immunity 25, 343–348 (2006).
Schwarz, D. S. et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115, 199–208 (2003).
Khvorova, A., Reynolds, A. & Jayasena, S. D. Functional siRNAs and miRNAs exhibit strand bias. Cell 115, 209–216 (2003).
Aza-Blanc, P. et al. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol. Cell 12, 627–637 (2003).
Liu, Q. et al. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301, 1921–1925 (2003). This paper established the paradigm that the RNase III proteins that act in RNA silencing pathways have dsRNA-binding protein partners, and showed that the Dicer partner R2D2 facilitates loading of the double-stranded siRNA into an Argonaute protein.
Tomari, Y., Matranga, C., Haley, B., Martinez, N. & Zamore, P. D. A protein sensor for siRNA asymmetry. Science 306, 1377–1380 (2004).
Matranga, C., Tomari, Y., Shin, C., Bartel, D. P. & Zamore, P. D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123, 607–620 (2005).
Kim, K., Lee, Y. S. & Carthew, R. W. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA 13, 22–29 (2006).
Leuschner, P. J., Ameres, S. L., Kueng, S. & Martinez, J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 7, 314–320 (2006).
Miyoshi, K., Tsukumo, H., Nagami, T., Siomi, H. & Siomi, M. C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 19, 2837–2848 (2005).
Rand, T. A., Petersen, S., Du, F. & Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123, 621–629 (2005).
Horwich, M. D. et al. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol. 17, 1265–1272 (2007).
Pelisson, A., Sarot, E., Payen-Groschene, G. & Bucheton, A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J. Virol. 81, 1951–1960 (2007). The discovery that siRNAs are 2′- O -methylated in flies.
Yu, B. et al. Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935 (2005). The first report of small silencing RNA methylation. This paper alerted the field to the idea that small RNA stability and function could be controlled by post-transcriptional modification.
Ramachandran, V. & Chen, X. Small RNA metabolism in Arabidopsis. Trends Plant Sci. 13, 368–374 (2008).
Li, J., Yang, Z., Yu, B., Liu, J. & Chen, X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15, 1501–1507 (2005).
Tolia, N. H. & Joshua-Tor, L. Slicer and the Argonautes. Nature Chem. Biol. 3, 36–43 (2007).
Hamilton, A., Voinnet, O., Chappell, L. & Baulcombe, D. Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671–4679 (2002).
Tang, G., Reinhart, B. J., Bartel, D. P. & Zamore, P. D. A biochemical framework for RNA silencing in plants. Genes Dev. 17, 49–63 (2003).
Dunoyer, P., Himber, C., Ruiz-Ferrer, V., Alioua, A. & Voinnet, O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nature Genet. 39, 848–856 (2007).
Bouche, N., Lauressergues, D., Gasciolli, V. & Vaucheret, H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25, 3347–3356 (2006).
Deleris, A. et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science 313, 68–71 (2006).
Gasciolli, V., Mallory, A. C., Bartel, D. P. & Vaucheret, H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 15, 1494–1500 (2005).
Xie, Z. et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2, E104 (2004).
Chan, S. W. Inputs and outputs for chromatin-targeted RNAi. Trends Plant Sci. 13, 383–389 (2008).
Qu, F. et al. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 79, 15209–15217 (2005).
Voinnet, O. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 13, 317–328 (2008).
Gazzani, S., Lawrenson, T., Woodward, C., Headon, D. & Sablowski, R. A link between mRNA turnover and RNA interference in Arabidopsis. Science 306, 1046–1048 (2004).
Gy, I. et al. Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors. Plant Cell 19, 3451–3461 (2007).
Ambros, V., Lee, R. C., Lavanway, A., Williams, P. T. & Jewell, D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 13, 807–818 (2003).
Zilberman, D., Cao, X. & Jacobsen, S. E. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299, 716–719 (2003).
Chan, S. W. et al. RNA silencing genes control de novo DNA methylation. Science 303, 1336 (2004).
Zheng, X., Zhu, J., Kapoor, A. & Zhu, J. K. Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 26, 1691–1701 (2007).
Boutet, S. et al. Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 13, 843–848 (2003).
El-Shami, M. et al. Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev. 21, 2539–2544 (2007).
Herr, A. J., Jensen, M. B., Dalmay, T. & Baulcombe, D. C. RNA polymerase IV directs silencing of endogenous DNA. Science 308, 118–120 (2005).
Kanoh, J., Sadaie, M., Urano, T. & Ishikawa, F. Telomere binding protein Taz1 establishes Swi6 heterochromatin independently of RNAi at telomeres. Curr. Biol. 15, 1808–1819 (2005).
Lee, S. K. et al. Lentiviral delivery of short hairpin RNAs protects CD4 T cells from multiple clades and primary isolates of HIV. Blood 106, 818–826 (2005).
Onodera, Y. et al. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120, 613–622 (2005).
Pontier, D. et al. Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev. 19, 2030–2040 (2005).
Tran, R. K. et al. DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr. Biol. 15, 154–159 (2005).
Llave, C., Kasschau, K. D., Rector, M. A. & Carrington, J. C. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14, 1605–1619 (2002).
Mette, M. F., Aufsatz, W., van der Winden, J., Matzke, M. A. & Matzke, A. J. Transcriptional silencing and promoter methylation triggered by double-stranded RNA. EMBO J. 19, 5194–5201 (2000).
Vazquez, F. et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16, 69–79 (2004).
Peragine, A., Yoshikawa, M., Wu, G., Albrecht, H. L. & Poethig, R. S. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 18, 2368–2379 (2004).
Yoshikawa, M., Peragine, A., Park, M. Y. & Poethig, R. S. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19, 2164–2175 (2005).
Williams, L., Carles, C. C., Osmont, K. S. & Fletcher, J. C. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl Acad. Sci. USA 102, 9703–9708 (2005).
Allen, E., Xie, Z., Gustafson, A. M. & Carrington, J. C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221 (2005).
Montgomery, T. A. et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133, 128–141 (2008).
Axtell, M. J., Jan, C., Rajagopalan, R. & Bartel, D. P. A two-hit trigger for siRNA biogenesis in plants. Cell 127, 565–577 (2006).
Katiyar-Agarwal, S. et al. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl Acad. Sci. USA 103, 18002–18007 (2006).
Borsani, O., Zhu, J., Verslues, P. E., Sunkar, R. & Zhu, J. K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123, 1279–1291 (2005).
Zhang, D. & Trudeau, V. L. The XS domain of a plant specific SGS3 protein adopts a unique RNA recognition motif (RRM) fold. Cell Cycle 7, 2268–2270 (2008).
Katiyar-Agarwal, S., Gao, S., Vivian-Smith, A. & Jin, H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 21, 3123–3134 (2007).
Yang, N. & Kazazian, H. H. J. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nature Struct. Mol. Biol. 13, 763–771 (2006).
Ghildiyal, M. et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320, 1077–1081 (2008).
Czech, B. et al. An endogenous small interfering RNA pathway in Drosophila. Nature 453, 798–802 (2008).
Okamura, K. et al. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature 453, 803–806 (2008).
Kawamura, Y. et al. Drosophila endogenous small RNAs bind to Argonaute 2 in somatic cells. Nature 453, 793–797 (2008).
Okamura, K., Balla, S., Martin, R., Liu, N. & Lai, E. C. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nature Struct. Mol. Biol. 15, 581–590 (2008).
Chung, W. J., Okamura, K., Martin, R. & Lai, E. C. Endogenous RNA interference provides a somatic defense against Drosophila transposons. Curr. Biol. 18, 795–802 (2008).
Tabara, H. et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132 (1999). The first study to link an Argonaute protein to an RNA silencing pathway. The authors showed that the Argonaute protein RDE-1 is required for RNAi in worms.
Ketting, R. F., Haverkamp, T. H., van Luenen, H. G. & Plasterk, R. H. mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99, 133–141 (1999).
Zhou, R. et al. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol. Cell 32, 592–599 (2008).
Förstemann, K. et al. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 3, e236 (2005).
Tam, O. H. et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453, 534–538 (2008).
Watanabe, T. et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453, 539–543 (2008).
Sasidharan, R. & Gerstein, M. Genomics: protein fossils live on as RNA. Nature 453, 729–731 (2008).
Neilson, J. R. & Sharp, P. A. Small RNA regulators of gene expression. Cell 134, 899–902 (2008).
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993). This paper reported the discovery of the first miRNA, lin-4 , and, together with reference 92, proposed that it regulated an mRNA, lin-14 , by partially base-pairing to sites in the 3′ UTR, a relationship now considered as the hallmark of miRNA regulation.
Wightman, B., Burglin, T. R., Gatto, J., Arasu, P. & Ruvkun, G. Negative regulatory sequences in the lin-14 3′-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 5, 1813–1124 (1991).
Wightman, B., Ha, I. & Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 (1993).
Lagos-Quintana, M., Rauhut, R., Lendeckel, W. & Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 294, 853–858 (2001).
Lee, R. C. & Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862–864 (2001).
Lau, N. C., Lim, L. P., Weinstein, E. G. & Bartel, D. P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858–862 (2001). References 93–95 ushered in the age of miRNA discovery, increasing the number of known miRNAs from 2 to 76.
Griffiths-Jones, S., Saini, H. K., van Dongen, S. & Enright, A. J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 36, D154–D158 (2008).
Lee, Y., Jeon, K., Lee, J. T., Kim, S. & Kim, V. N. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 21, 4663–4670 (2002).
Lee, Y. et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 23, 4051–4060 (2004).
Cai, X., Hagedorn, C. H. & Cullen, B. R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10, 1957–1966 (2004).
Birney, E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007).
Lee, Y. et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419 (2003). The discovery that Drosha generates pre-miRNAs in the nucleus from longer, primary transcripts.
Denli, A. M., Tops, B. B., Plasterk, R. H., Ketting, R. F. & Hannon, G. J. Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235 (2004).
Gregory, R. I. et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240 (2004).
Han, J. et al. The Drosha–DGCR8 complex in primary microRNA processing. Genes Dev. 18, 3016–3027 (2004).
Landthaler, M., Yalcin, A. & Tuschl, T. The human DiGeorge syndrome critical region gene 8 and its D. melanogaster homolog are required for miRNA biogenesis. Curr. Biol. 14, 2162–2167 (2004).
Yi, R., Qin, Y., Macara, I. G. & Cullen, B. R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17, 3011–3016 (2003).
Bohnsack, M. T., Czaplinski, K. & Gorlich, D. Exportin 5 is a Ran GTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10, 185–191 (2004).
Lund, E., Guttinger, S., Calado, A., Dahlberg, J. E. & Kutay, U. Nuclear export of microRNA precursors. Science 303, 95–98 (2004).
Zeng, Y. & Cullen, B. R. Structural requirements for pre-microRNA binding and nuclear export by Exportin 5. Nucleic Acids Res. 32, 4776–4785 (2004).
Chendrimada, T. P. et al. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436, 740–744 (2005).
Jiang, F. et al. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 19, 1674–1679 (2005).
Lee, Y. et al. The role of PACT in the RNA silencing pathway. EMBO J. 25, 522–532 (2006).
Saito, K., Ishizuka, A., Siomi, H. & Siomi, M. C. Processing of pre-microRNAs by the Dicer-1–Loquacious complex in Drosophila cells. PLoS Biol. 3, e235 (2005).
Ruby, J. G. et al. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell 127, 1193–1207 (2006).
Okamura, K., Hagen, J. W., Duan, H., Tyler, D. M. & Lai, E. C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130, 89–100 (2007).
Ruby, J. G., Jan, C. H. & Bartel, D. P. Intronic microRNA precursors that bypass Drosha processing. Nature 448, 83–86 (2007).
Glazov, E. A. et al. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res. 18, 957–964 (2008).
Berezikov, E., Chung, W. J., Willis, J., Cuppen, E. & Lai, E. C. Mammalian mirtron genes. Mol. Cell 28, 328–336 (2007).
Babiarz, J. E., Ruby, J. G., Wang, Y., Bartel, D. P. & Blelloch, R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 22, 2773–2785 (2008).
Park, W., Li, J., Song, R., Messing, J. & Chen, X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12, 1484–1495 (2002).
Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B. & Bartel, D. P. MicroRNAs in plants. Genes Dev. 16, 1616–1626 (2002).
Papp, I. et al. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 132, 1382–1390 (2003).
Vazquez, F., Gasciolli, V., Crete, P. & Vaucheret, H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14, 346–351 (2004).
Park, M. Y., Wu, G., Gonzalez-Sulser, A., Vaucheret, H. & Poethig, R. S. Nuclear processing and export of microRNAs in Arabidopsis. Proc. Natl Acad. Sci. USA 102, 3691–3696 (2005).
Yang, Z., Ebright, Y. W., Yu, B. & Chen, X. HEN1 recognizes 21–24 nt small RNA duplexes and deposits a methyl group onto the 2′ OH of the 3′ terminal nucleotide. Nucleic Acids Res. 34, 667–675 (2006).
Hutvágner, G. & Zamore, P. D. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060 (2002).
Meister, G. et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 15, 185–197 (2004).
Liu, J. et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441 (2004).
Mansfield, J. H. et al. MicroRNA-responsive 'sensor' transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nature Genet. 36, 1079–1083 (2004).
Pfeffer, S. et al. Identification of virus-encoded microRNAs. Science 304, 734–736 (2004).
Yekta, S., Shih, I. H. & Bartel, D. P. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304, 594–596 (2004).
Davis, E. et al. RNAi-mediated allelic trans-interaction at the imprinted Rtl1/Peg11 locus. Curr. Biol. 15, 743–749 (2005).
Sullivan, C. S., Grundhoff, A. T., Tevethia, S., Pipas, J. M. & Ganem, D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435, 682–686 (2005).
Rhoades, M. W. et al. Prediction of plant microRNA targets. Cell 110, 513–520 (2002).
Brennecke, J., Stark, A., Russell, R. B. & Cohen, S. M. Principles of microRNA-target recognition. PLoS Biol. 3, e85 (2005).
Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P. & Burge, C. B. Prediction of mammalian microRNA targets. Cell 115, 787–798 (2003). This paper reports the first computational evidence for a seed sequence and uses seed pairing to identify miRNA targets.
Lewis, B. P., Burge, C. B. & Bartel, D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
Stark, A., Brennecke, J., Bushati, N., Russell, R. B. & Cohen, S. M. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123, 1133–1146 (2005).
Xie, X. et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434, 338–345 (2005).
Lai, E. C. Predicting and validating microRNA targets. Genome Biol. 5, 115 (2004).
Ameres, S. L., Martinez, J. & Schroeder, R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 130, 101–112 (2007).
Haley, B. & Zamore, P. D. Kinetic analysis of the RNAi enzyme complex. Nature Struct. Mol. Biol. 11, 599–606 (2004).
Baek, D. et al. The impact of microRNAs on protein output. Nature 455, 64–71 (2008).
Selbach, M. et al. Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 (2008).
Kong, Y. W. et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc. Natl Acad. Sci. USA 105, 8866–8871 (2008).
Brodersen, P. et al. Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190 (2008).
Stefani, G. & Slack, F. J. Small non-coding RNAs in animal development. Nature Rev. Mol. Cell Biol. 9, 219–230 (2008).
Yekta, S., Tabin, C. J. & Bartel, D. P. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nature Rev. Genet. 9, 789–796 (2008).
Bartel, D. P. & Chen, C. Z. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nature Rev. Genet. 5, 396–400 (2004).
Bernstein, E. et al. Dicer is essential for mouse development. Nature Genet. 35, 215–217 (2003).
Wienholds, E. et al. MicroRNA expression in zebrafish embryonic development. Science 309, 310–311 (2005).
Giraldez, A. J. et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science 308, 833–838 (2005).
Hatfield, S. D. et al. Stem cell division is regulated by the microRNA pathway. Nature 435, 974–978 (2005).
Mudhasani, R. et al. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J. Cell Biol. 181, 1055–1063 (2008).
Landgraf, P. et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414 (2007).
Wienholds, E. & Plasterk, R. H. MicroRNA function in animal development. FEBS Lett. 579, 5911–5922 (2005).
Aravin, A. A. et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11, 1017–1027 (2001). This paper was the first to identify piRNAs, then called repeat-associated siRNAs.
Vagin, V. V. et al. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324 (2006). The first report that piRNAs are distinct from siRNAs. This paper proposed that piRNAs are made from single-stranded precursors without the involvement of Dicer, are 3′ modified and are bound to Piwi proteins, and that they are largely antisense to selfish genetic elements.
Saito, K. et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 20, 2214–2222 (2006).
Saito, K. et al. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 21, 1603–1608 (2007).
Kirino, Y. & Mourelatos, Z. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nature Struct. Mol. Biol. 14, 347–348 (2007).
Ohara, T. et al. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nature Struct. Mol. Biol. 14, 349–350 (2007).
Houwing, S. et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129, 69–82 (2007).
Aravin, A. et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207 (2006).
Lau, N. C. et al. Characterization of the piRNA complex from rat testes. Science 313, 363–367 (2006).
Aravin, A. A., Sachidanandam, R., Girard, A., Fejes-Toth, K. & Hannon, G. J. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747 (2007).
Girard, A., Sachidanandam, R., Hannon, G. J. & Carmell, M. A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202 (2006).
Grivna, S. T., Beyret, E., Wang, Z. & Lin, H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20, 1709–1714 (2006).
Grivna, S. T., Pyhtila, B. & Lin, H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc. Natl Acad. Sci. USA 103, 13415–13420 (2006).
Kato, Y. et al. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum. Mol. Genet. 16, 2272–2280 (2007).
Lees-Murdock, D. J., De Felici, M. & Walsh, C. P. Methylation dynamics of repetitive DNA elements in the mouse germ cell lineage. Genomics 82, 230–237 (2003).
Aravin, A. A. et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 (2008).
Batista, P. J. et al. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31, 67–78 (2008).
Das, P. P. et al. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 31, 79–90 (2008).
Wang, G. & Reinke, V. A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 18, 861–867 (2008).
Ruby, J. G. et al. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 17, 1850–1864 (2007).
Brennecke, J. et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128, 1089–1103 (2007). This paper, along with reference 184, proposed the 'ping-pong' model for piRNA biogenesis. The authors were the first to use high-throughput sequencing data to deduce the biogenesis mechanism of a small RNA pathway.
Nishida, K. M. et al. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13, 1911–1922 (2007).
Prud'homme, N., Gans, M., Masson, M., Terzian, C. & Bucheton, A. flamenco, a gene controlling the gypsy retrovirus of Drosophila melanogaster. Genetics 139, 697–711 (1995).
Pelisson, A. et al. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 13, 4401–4411 (1994).
Desset, S., Buchon, N., Meignin, C., Coiffet, M. & Vaury, C. In Drosophila melanogaster the COM locus directs the somatic silencing of two retrotransposons through both Piwi-dependent and -independent pathways. PLoS ONE 3, e1526 (2008).
Desset, S., Meignin, C., Dastugue, B. & Vaury, C. COM, a heterochromatic locus governing the control of independent endogenous retroviruses from Drosophila melanogaster. Genetics 164, 501–509 (2003).
Mevel-Ninio, M. T., Pelisson, A., Kinder, J., Campos, A. R. & Bucheton, A. The flamenco locus controls the gypsy and ZAM retroviruses and is required for Drosophila oogenesis. Genetics 175, 1615–24 (2007).
Gunawardane, L. S. et al. A Slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315, 1587–1590 (2007).
Cox, D. N. et al. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12, 3715–3727 (1998).
Cox, D. N., Chao, A. & Lin, H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development 127, 503–514 (2000).
Aravin, A. A. et al. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell Biol. 24, 6742–6750 (2004).
Schupbach, T. & Wieschaus, E. Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129, 1119–1136 (1991).
Klattenhoff, C. et al. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell 12, 45–55 (2007).
Pal-Bhadra, M. et al. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303, 669–672 (2004).
Tomari, Y., Du, T. & Zamore, P. D. Sorting of Drosophila small silencing RNAs. Cell 130, 299–308 (2007).
Förstemann, K., Horwich, M. D., Wee, L.-M., Tomari, Y. & Zamore, P. D. Drosophila microRNAs are sorted into functionally distinct Argonaute protein complexes after their production by Dicer-1. Cell 130, 287–297 (2007).
Mi, S. et al. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127 (2008).
Vaucheret, H. Plant ARGONAUTES. Trends Plant Sci. 13, 350–358 (2008).
Tokumaru, S., Suzuki, M., Yamada, H., Nagino, M. & Takahashi, T. let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis 29, 2073–2077 (2008).
Forman, J. J., Legesse-Miller, A. & Coller, H. A. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl Acad. Sci. USA 105, 14879–14884 (2008).
Xie, Z., Kasschau, K. D. & Carrington, J. C. Negative feedback regulation of Dicer-like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr. Biol. 13, 784–789 (2003).
Vaucheret, H., Vazquez, F., Crete, P. & Bartel, D. P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197 (2004).
Vaucheret, H., Mallory, A. C. & Bartel, D. P. AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol. Cell 22, 129–136 (2006).
Zhao, T. et al. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 21, 1190–1203 (2007).
Molnar, A., Schwach, F., Studholme, D. J., Thuenemann, E. C. & Baulcombe, D. C. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature 447, 1126–1129 (2007).
Grimson, A. et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature 455, 1193–1197 (2008). This paper established that miRNAs and piRNAs existed before the divergence of bilaterian animals from the animal kingdom.
Brennecke, J. et al. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322, 1387–1392 (2008).
Sijen, T. et al. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107, 465–476 (2001).
Voinnet, O., Vain, P., Angell, S. & Baulcombe, D. C. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187 (1998).
Vaistij, F. E., Jones, L. & Baulcombe, D. C. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14, 857–867 (2002).
Fusaro, A. F. et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 7, 1168–1175 (2006).
Moissiard, G., Parizotto, E. A., Himber, C. & Voinnet, O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA 13, 1268–1278 (2007).
Sijen, T., Steiner, F. A., Thijssen, K. L. & Plasterk, R. H. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315, 244–247 (2007).
Pak, J. & Fire, A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science 315, 241–244 (2007).
Smardon, A. et al. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10, 169–178 (2000).
Aoki, K., Moriguchi, H., Yoshioka, T., Okawa, K. & Tabara, H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26, 5007–5019 (2007).
Makeyev, E. V. & Bamford, D. H. Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol. Cell 10, 1417–1427 (2002).
Tijsterman, M., Ketting, R. F., Okihara, K. L., Sijen, T. & Plasterk, R. H. RNA helicase MUT-14-dependent gene silencing triggered in C. elegans by short antisense RNAs. Science 295, 694–697 (2002).
Caplen, N. J. et al. Rescue of polyglutamine-mediated cytotoxicity by double-stranded RNA-mediated RNA interference. Hum. Mol. Genet. 11, 175–184 (2002).
Xia, H., Mao, Q., Paulson, H. L. & Davidson, B. L. siRNA-mediated gene silencing in vitro and in vivo. Nature Biotechnol. 20, 1006–1010 (2002).
Ding, H. et al. Selective silencing by RNAi of a dominant allele that causes amyotrophic lateral sclerosis. Aging Cell 2, 209–217 (2003).
Miller, V. M. et al. Allele-specific silencing of dominant disease genes. Proc. Natl Acad. Sci. USA 100, 7195–7200 (2003).
Martinez, L. A. et al. Synthetic small inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore p53 pathways. Proc. Natl Acad. Sci. USA 99, 14849–14854 (2002).
Schwarz, D. S., Hutvagner, G., Haley, B. & Zamore, P. D. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10, 537–548 (2002).
Nykanen, A., Haley, B. & Zamore, P. D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107, 309–321 (2001).
Martinez, J., Patkaniowska, A., Urlaub, H., Lührmann, R. & Tuschl, T. Single stranded antisense siRNA guide target RNA cleavage in RNAi. Cell 110, 563–574 (2002).
Roignant, J. Y. et al. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA 9, 299–308 (2003).
Rusk, N. & Kiermer, V. Primer: Sequencing — the next generation. Nature Methods 5, 15 (2008).
Johnston, R. J. & Hobert, O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426, 845–849 (2003).
Jones-Rhoades, M. W. & Bartel, D. P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 14, 787–799 (2004).
Elbashir, S. M. et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498 (2001). The breakthrough paper that showed siRNAs could be used in cultured somatic mammalian cells to phenocopy loss-of-function genetic mutations.
Sijen, T. & Plasterk, R. H. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426, 310–314 (2003).
Acknowledgements
We thank H. Seitz for bioinformatic assistance and X. Chen, I. Pekker and S. Ameres for helpful discussions. This work was supported, in part, by grants from the National Institutes of Health to P.D.Z. (GM065236 and GM062862).
Author information
Authors and Affiliations
Corresponding author
Related links
Glossary
- Argonaute
-
Argonaute proteins are the effectors of small RNA-directed silencing. Small RNAs guide Argonautes to their RNA targets; Argonautes carry out the regulation. Argonaute proteins are characterized by two domains — Piwi (a ribonuclease domain) and PAZ (a ssRNA-binding module).
- Retrotransposon
-
A transposable element that replicates via an RNA intermediate, which is converted by reverse transcriptase to cDNA. The cDNA can be inserted into genomic DNA, increasing the number of copies of the retrotransposon in the genome.
- Sequencing depth
-
The number of sequences obtained by high-throughput sequencing for a specific sample. Sequencing depth is often expressed as 'genome-matching reads'.
- Pachytene
-
Pachytene is the stage of meiotic prophase during which the homologous chromosomes condense and separate into two chromatids. At the pachytene stage, homologous chromosomes pair and crossing over — the exchange of DNA segments — occurs between two non-sister chromatids.
Rights and permissions
About this article
Cite this article
Ghildiyal, M., Zamore, P. Small silencing RNAs: an expanding universe. Nat Rev Genet 10, 94–108 (2009). https://doi.org/10.1038/nrg2504
Issue Date:
DOI: https://doi.org/10.1038/nrg2504
This article is cited by
-
Silencing of ApCht7 and ApCht10 revealed their function and evaluation of their potential as RNAi targets in Acyrthosiphon pisum
Journal of Pest Science (2024)
-
MicroRNA cross-talk between Monilinia fungal pathogens and peach host
Phytoparasitica (2024)
-
The emerging role of the piRNA/PIWI complex in respiratory tract diseases
Respiratory Research (2023)
-
Small RNAs, spermatogenesis, and male infertility: a decade of retrospect
Reproductive Biology and Endocrinology (2023)
-
Small RNA sequencing of field Culex mosquitoes identifies patterns of viral infection and the mosquito immune response
Scientific Reports (2023)