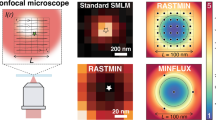
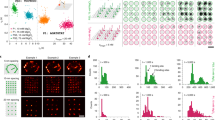
Abstract
Validating and testing a fluorescence microscope or a microscopy method requires defined samples that can be used as standards. DNA origami is a new tool that provides a framework to place defined numbers of small molecules such as fluorescent dyes or proteins in a programmed geometry with nanometer precision. The flexibility and versatility in the design of DNA origami microscopy standards makes them ideally suited for the broad variety of emerging super-resolution microscopy methods. As DNA origami structures are durable and portable, they can become a universally available specimen to check the everyday functionality of a microscope. The standards are immobilized on a glass slide, and they can be imaged without further preparation and can be stored for up to 6 months. We describe a detailed protocol for the design, production and use of DNA origami microscopy standards, and we introduce a DNA origami rectangle, bundles and a nanopillar as fluorescent nanoscopic rulers. The protocol provides procedures for the design and realization of fluorescent marks on DNA origami structures, their production and purification, quality control, handling, immobilization, measurement and data analysis. The procedure can be completed in 1–2 d.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout












Similar content being viewed by others
References
Rothemund, P.W. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Douglas, S.M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Ke, Y., Ong, L.L., Shih, W.M. & Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 338, 1177–1183 (2012).
Douglas, S.M., Bachelet, I. & Church, G.M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012).
Chen, Z., Lan, X. & Wang, Q. DNA origami directed large-scale fabrication of nanostructures resembling room temperature single-electron transistors. Small 9, 3567–3571 (2013).
Kershner, R.J. et al. Placement and orientation of individual DNA shapes on lithographically patterned surfaces. Nat. Nanotechnol. 4, 557–561 (2009).
Langecker, M. et al. Synthetic lipid membrane channels formed by designed DNA nanostructures. Science 338, 932–936 (2012).
Acuna, G.P. et al. Fluorescence enhancement at docking sites of DNA-directed self-assembled nanoantennas. Science 338, 506–510 (2012).
Torring, T., Voigt, N.V., Nangreave, J., Yan, H. & Gothelf, K.V. DNA origami: a quantum leap for self-assembly of complex structures. Chem. Soc. Rev. 40, 5636–5646 (2011).
Rajendran, A., Endo, M. & Sugiyama, H. Single-molecule analysis using DNA origami. Angew. Chem. Int. Ed. Engl. 51, 874–890 (2012).
Pinheiro, A.V., Han, D., Shih, W.M. & Yan, H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 6, 763–772 (2011).
Wang, Z.G., Song, C. & Ding, B. Functional DNA nanostructures for photonic and biomedical applications. Small 9, 2210–2222 (2013).
Bellot, G., McClintock, M.A., Chou, J.J. & Shih, W.M. DNA nanotubes for NMR structure determination of membrane proteins. Nat. Protoc. 8, 755–770 (2013).
Kim, D.N., Kilchherr, F., Dietz, H. & Bathe, M. Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 40, 2862–2868 (2012).
Castro, C.E. et al. A primer to scaffolded DNA origami. Nat. Methods 8, 221–229 (2011).
Steinhauer, C., Jungmann, R., Sobey, T., Simmel, F. & Tinnefeld, P. DNA origami as a nanoscopic ruler for super-resolution microscopy. Angew. Chem. Int. Ed. Engl. 48, 8870–8873 (2009).
Schmied, J.J. et al. DNA origami nanopillars as standards for three-dimensional super-resolution microscopy. Nano Lett. 13, 781–785 (2013).
Schmied, J.J. et al. Fluorescence and super-resolution standards based on DNA origami. Nat. Methods 9, 1133–1134 (2012).
Hell, S.W. Far-field optical nanoscopy. Science 316, 1153–1158 (2007).
Huang, B., Bates, M. & Zhuang, X. Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 78, 993–1016 (2009).
Rust, M.J., Bates, M. & Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 (2006).
van de Linde, S., Heilemann, M. & Sauer, M. Live-cell super-resolution imaging with synthetic fluorophores. Annu. Rev. Phys. Chem. 63, 519–540 (2012).
Klar, T.A., Jakobs, S., Dyba, M., Egner, A. & Hell, S.W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. USA 97, 8206–8210 (2000).
Willig, K.I., Harke, B., Medda, R. & Hell, S.W. STED microscopy with continuous wave beams. Nat. Methods 4, 915–918 (2007).
Vicidomini, G. et al. Sharper low-power STED nanoscopy by time gating. Nat. Methods 8, 571–573 (2011).
Harke, B., Ullal, C.K., Keller, J. & Hell, S.W. Three-dimensional nanoscopy of colloidal crystals. Nano Lett. 8, 1309–1313 (2008).
Gustafsson, M.G. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82–87 (2000).
Gustafsson, M.G. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc. Natl. Acad. Sci. USA 102, 13081–13086 (2005).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Hess, S.T., Girirajan, T.P. & Mason, M.D. Ultra-high-resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258–4272 (2006).
Heilemann, M. et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47, 6172–6176 (2008).
Folling, J. et al. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat. Methods 5, 943–945 (2008).
Steinhauer, C., Forthmann, C., Vogelsang, J. & Tinnefeld, P. Superresolution microscopy on the basis of engineered dark states. J. Am. Chem. Soc. 130, 16840–16841 (2008).
Thompson, M.A., Casolari, J.M., Badieirostami, M., Brown, P.O. & Moerner, W.E. Three-dimensional tracking of single mRNA particles in Saccharomyces cerevisiae using a double-helix point spread function. Proc. Natl. Acad. Sci. USA 107, 17864–17871 (2010).
Huang, B., Wang, W., Bates, M. & Zhuang, X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319, 810–813 (2008).
Juette, M.F. et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat. Methods 5, 527–529 (2008).
Dertinger, T., Colyer, R., Iyer, G., Weiss, S. & Enderlein, J. Fast, background-free, 3D super-resolution optical fluctuation imaging (SOFI). Proc. Natl. Acad. Sci. USA 106, 22287–22292 (2009).
Xu, K., Zhong, G. & Zhuang, X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science 339, 452–456 (2013).
Chojnacki, J. et al. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 338, 524–528 (2012).
Szymborska, A. et al. Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science 341, 655–658 (2013).
Bates, M., Huang, B. & Zhuang, X. Super-resolution microscopy by nanoscale localization of photo-switchable fluorescent probes. Curr. Opin. Chem. Biol. 12, 505–514 (2008).
Ram, S., Ward, E.S. & Ober, R.J. Beyond Rayleigh's criterion: a resolution measure with application to single-molecule microscopy. Proc. Natl. Acad. Sci. USA 103, 4457–4462 (2006).
Hell, S.W. Strategy for far-field optical imaging and writing without diffraction limit. Phys. Lett. A 326, 140–145 (2004).
Fitzgerald, J.E., Lu, J. & Schnitzer, M.J. Estimation theoretic measure of resolution for stochastic localization microscopy. Phys. Rev. Lett. 109, 048102 (2012).
Nieuwenhuizen, R.P. et al. Measuring image resolution in optical nanoscopy. Nat. Methods 10, 557–562 (2013).
Shroff, H., Galbraith, C.G., Galbraith, J.A. & Betzig, E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 5, 417–423 (2008).
Vogelsang, J. et al. Make them blink: probes for super-resolution microscopy. ChemPhysChem 11, 2475–2490 (2010).
Bellec, M. et al. 3D patterning at the nanoscale of fluorescent emitters in glass. J. Phys. Chem. C 114, 15584–15588 (2010).
Cordes, T. et al. Resolving single-molecule assembled patterns with super-resolution blink-microscopy. Nano Lett. 10, 645–651 (2010).
Whitesides, G.M., Mathias, J.P. & Seto, C.T. Molecular self-assembly and nanochemistry: a chemical strategy for the synthesis of nanostructures. Science 254, 1312–1319 (1991).
Seeman, N.C. Nanomaterials based on DNA. Annu. Rev. Biochem. 79, 65–87 (2010).
Loschberger, A. et al. Super-resolution imaging visualizes the eightfold symmetry of gp210 proteins around the nuclear pore complex and resolves the central channel with nanometer resolution. J. Cell Sci. 125, 570–575 (2012).
Gottfert, F. et al. Coaligned dual-channel STED nanoscopy and molecular diffusion analysis at 20 nm resolution. Biophys. J. 105, L01–L03 (2013).
Jungmann, R., Liedl, T., Sobey, T.L., Shih, W. & Simmel, F.C. Isothermal assembly of DNA origami structures using denaturing agents. J. Am. Chem. Soc. 130, 10062–10063 (2008).
Kauert, D.J., Kurth, T., Liedl, T. & Seidel, R. Direct mechanical measurements reveal the material properties of three-dimensional DNA origami. Nano Lett. 11, 5558–5563 (2011).
Stein, I.H., Schuller, V., Bohm, P., Tinnefeld, P. & Liedl, T. Single-molecule FRET ruler based on rigid DNA origami blocks. ChemPhysChem 12, 689–695 (2011).
Lin, C. et al. Submicrometre geometrically encoded fluorescent barcodes self-assembled from DNA. Nat. Chem. 4, 832–839 (2012).
Zhuang, X. et al. Fluorescence quenching: A tool for single-molecule protein-folding study. Proc. Natl. Acad. Sci. USA 97, 14241–14244 (2000).
Mei, Q. et al. Stability of DNA origami nanoarrays in cell lysate. Nano Lett. 11, 1477–1482 (2011).
Gietl, A., Holzmeister, P., Grohmann, D. & Tinnefeld, P. DNA origami as biocompatible surface to match single-molecule and ensemble experiments. Nucleic Acids Res. 40, e110 (2012).
Rajendran, A., Endo, M., Katsuda, Y., Hidaka, K. & Sugiyama, H. Photo-cross-linking-assisted thermal stability of DNA origami structures and its application for higher-temperature self-assembly. J. Am. Chem. Soc. 133, 14488–14491 (2011).
van de Linde, S. et al. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 6, 991–1009 (2011).
Schulz, O. et al. Tip induced fluorescence quenching for nanometer optical and topographical resolution. Opt. Nanosc. 2, 1 (2013).
Kurz, A. et al. Counting fluorescent dye molecules on DNA origami by means of photon statistics. Small 9, 4061–4068 (2013).
Woo, S. & Rothemund, P.W. Programmable molecular recognition based on the geometry of DNA nanostructures. Nat. Chem. 3, 620–627 (2011).
Li, Z., Wang, L., Yan, H. & Liu, Y. Effect of DNA hairpin loops on the twist of planar DNA origami tiles. Langmuir 28, 1959–1965 (2012).
Hein, B., Willig, K.I. & Hell, S.W. Stimulated emission depletion (STED) nanoscopy of a fluorescent protein-labeled organelle inside a living cell. Proc. Natl. Acad. Sci. USA 105, 14271–14276 (2008).
Prabhat, P. et al. Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy. Proc. Natl. Acad. Sci. USA 104, 5889–5894 (2007).
Shtengel, G. et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc. Natl. Acad. Sci. USA 106, 3125–3130 (2009).
Pavani, S.R. et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc. Natl. Acad. Sci. USA 106, 2995–2999 (2009).
Baddeley, D., Cannell, M.B. & Soeller, C. Three-dimensional sub-100 nm super-resolution imaging of biological samples using a phase ramp in the objective pupil. Nano Res. 4, 589–598 (2011).
Douglas, S.M. et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009).
Schreiber, R. et al. DNA origami-templated growth of arbitrarily shaped metal nanoparticles. Small 7, 1795–1799 (2011).
Derr, N.D. et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 338, 662–665 (2012).
Sobczak, J.P., Martin, T.G., Gerling, T. & Dietz, H. Rapid folding of DNA into nanoscale shapes at constant temperature. Science 338, 1458–1461 (2012).
Gould, T.J., Verkhusha, V.V. & Hess, S.T. Imaging biological structures with fluorescence photoactivation localization microscopy. Nat. Protoc. 4, 291–308 (2009).
Wolter, S. et al. rapidSTORM: accurate, fast open-source software for localization microscopy. Nat. Methods 9, 1040–1041 (2012).
Holden, S.J., Uphoff, S. & Kapanidis, A.N. DAOSTORM: an algorithm for high- density super-resolution microscopy. Nat. Methods 8, 279–280 (2011).
Henriques, R. et al. QuickPALM: 3D real-time photoactivation nanoscopy image processing in ImageJ. Nat. Methods 7, 339–340 (2010).
York, A.G., Ghitani, A., Vaziri, A., Davidson, M.W. & Shroff, H. Confined activation and subdiffractive localization enables whole-cell PALM with genetically expressed probes. Nat. Methods 8, 327–333 (2011).
Huang, F. et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nat. Methods 10, 653–658 (2013).
Dickson, R.M., Cubitt, A.B., Tsien, R.Y. & Moerner, W.E. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature 388, 355–358 (1997).
Shen, W., Zhong, H., Neff, D. & Norton, M.L. NTA directed protein nanopatterning on DNA Origami nanoconstructs. J. Am. Chem. Soc. 131, 6660–6661 (2009).
Zhao, Z., Liu, Y. & Yan, H. Organizing DNA origami tiles into larger structures using preformed scaffold frames. Nano Lett. 11, 2997–3002 (2011).
Liu, W., Zhong, H., Wang, R. & Seeman, N.C. Crystalline two-dimensional DNA-origami arrays. Angew. Chem. Int. Ed. Engl. 50, 264–267 (2011).
Yang, Y., Han, D., Nangreave, J., Liu, Y. & Yan, H. DNA origami with double-stranded DNA as a unified scaffold. ACS Nano 6, 8209–8215 (2012).
Zhang, H. et al. Folding super-sized DNA origami with scaffold strands from long-range PCR. Chem. Commun. 48, 6405–6407 (2012).
Smith, C.S., Joseph, N., Rieger, B. & Lidke, K.A. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat. Methods 7, 373–375 (2010).
El Beheiry, M. & Dahan, M. ViSP: representing single-particle localizations in three dimensions. Nat. Methods 10, 689–690 (2013).
Hogberg, B., Liedl, T. & Shih, W.M. Folding DNA origami from a double-stranded source of scaffold. J. Am. Chem. Soc. 131, 9154–9155 (2009).
Messing, J. New M13 vectors for cloning. Methods Enzymol. 101, 20–78 (1983).
Yanisch-Perron, C., Vieira, J. & Messing, J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33, 103–119 (1985).
Douglas, S.M., Chou, J.J. & Shih, W.M. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl. Acad. Sci. USA 104, 6644–6648 (2007).
Zhu, R., Li, X., Zhao, X.S. & Yu, A. Photophysical properties of Atto655 dye in the presence of guanosine and tryptophan in aqueous solution. J. Phys. Chem. B 115, 5001–5007 (2011).
Doose, S., Neuweiler, H. & Sauer, M. Fluorescence quenching by photoinduced electron transfer: a reporter for conformational dynamics of macromolecules. ChemPhysChem 10, 1389–1398 (2009).
Bai, X.C., Martin, T.G., Scheres, S.H. & Dietz, H. Cryo-EM structure of a 3D DNA-origami object. Proc. Natl. Acad. Sci. USA 109, 20012–20017 (2012).
Acknowledgements
We gratefully acknowledge STED measurements from F. Göttfert and SIM measurements from L. Schermelleh. We thank R. Jungmann for the caDNAno file for the NRO structure. This work was supported by a starting grant (SiMBA, ERC-2010-StG-20091118) of the European Research Council, the Biophotonics IV program of the Federal Ministry of Education and Research (BMBF, VDI) (13N11461) and the German Research Foundation (DFG Ti329/6-1).
Author information
Authors and Affiliations
Contributions
J.J.S. developed and troubleshot the technique; M.R. performed dSTORM measurements; C.F. developed and troubleshot the software and algorithms for data analysis; E.P. developed and troubleshot the AFM measurements; B.W. carried out confocal measurements; T.D. produced the scaffold strands and quality-checked the structures; P.T. conceived the study and supervised the projects. All authors contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
Technische Universität Braunschweig has filed German and US patent applications covering parts of the work described in this manuscript.
Supplementary information
Supplementary Methods
caDNAno files (ZIP 854 kb)
Rights and permissions
About this article
Cite this article
Schmied, J., Raab, M., Forthmann, C. et al. DNA origami–based standards for quantitative fluorescence microscopy. Nat Protoc 9, 1367–1391 (2014). https://doi.org/10.1038/nprot.2014.079
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.079
This article is cited by
-
Expansion-enhanced super-resolution radial fluctuations enable nanoscale molecular profiling of pathology specimens
Nature Nanotechnology (2023)
-
Nuclease resistance of DNA nanostructures
Nature Reviews Chemistry (2021)
-
3D test sample for the calibration and quality control of stimulated emission depletion (STED) and confocal microscopes
Communications Biology (2021)
-
DNA origami
Nature Reviews Methods Primers (2021)
-
Picosecond time-resolved photon antibunching measures nanoscale exciton motion and the true number of chromophores
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.