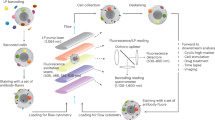
Abstract
We describe a flow-cytometry-based protocol for intracellular mRNA measurements in nonadherent mammalian cells using fluorescence in situ hybridization (FISH) probes. The method, which we call FISH-Flow, allows for high-throughput multiparametric measurements of gene expression, a task that was not feasible with earlier, microscopy-based approaches. The FISH-Flow protocol involves cell fixation, permeabilization and hybridization with a set of fluorescently labeled oligonucleotide probes. In this protocol, surface and intracellular protein markers can also be stained with fluorescently labeled antibodies for simultaneous protein and mRNA measurement. Moreover, a semiautomated, single-tube version of the protocol can be performed with a commercially available cell-wash device that reduces cell loss, operator time and interoperator variability. It takes ∼30 h to perform this protocol. An example of FISH-Flow measurements of cytokine mRNA induction by ex vivo stimulation of primed T cells with specific antigens is described.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Sanchez, A. & Golding, I. Genetic determinants and cellular constraints in noisy gene expression. Science 342, 1188–1193 (2013).
Femino, A.M., Fay, F.S., Fogarty, K. & Singer, R.H. Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998).
Raj, A., van den Bogaard, P., Rifkin, S.A., van Oudenaarden, A. & Tyagi, S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5, 877–879 (2008).
Rahman, S. & Zenklusen, D. Single-molecule resolution fluorescent in situ hybridization (smFISH) in the yeast S. cerevisiae. Methods Mol. Biol. 1042, 33–46 (2013).
Batish, M., Raj, A. & Tyagi, S. Single molecule imaging of RNA in situ. Methods Mol. Biol. 714, 3–13 (2011).
Tyagi, S. Imaging intracellular RNA distribution and dynamics in living cells. Nat. Methods 6, 331–338 (2009).
Bushkin, Y. et al. Profiling T cell activation using single-molecule fluorescence in situ hybridization and flow cytometry. J. Immunol. 194, 836–841 (2015).
Vir, P. et al. Single-cell cytokine gene expression in peripheral blood cells correlates with latent tuberculosis status. PLoS One 10, e0144904 (2015).
Chattopadhyay, P.K. & Roederer, M. Cytometry: today's technology and tomorrow's horizons. Methods 57, 251–258 (2012).
Hanley, M.B., Lomas, W., Mittar, D., Maino, V. & Park, E. Detection of low abundance RNA molecules in individual cells by flow cytometry. PLoS One 8, e57002 (2013).
Porichis, F. et al. Differential impact of PD-1 and/or interleukin-10 blockade on HIV-1-specific CD4 T cell and antigen-presenting cell functions. J. Virol. 88, 2508–2518 (2014).
Larsson, H.M. et al. Sorting live stem cells based on Sox2 mRNA expression. PLoS One 7, e49874 (2012).
McClellan, S. et al. mRNA detection in living cells: A next generation cancer stem cell identification technique. Methods 82, 47–54 (2015).
Perfetto, S.P., Chattopadhyay, P.K. & Roederer, M. Seventeen-colour flow cytometry: unravelling the immune system. Nat. Rev. Immunol. 4, 648–655 (2004).
Hulspas, R., O'Gorman, M.R., Wood, B.L., Gratama, J.W. & Sutherland, D.R. Considerations for the control of background fluorescence in clinical flow cytometry. Cytometry B Clin. Cytom. 76, 355–364 (2009).
Perfetto, S.P. et al. Amine-reactive dyes for dead cell discrimination in fixed samples. Curr. Protoc. Cytom. Chapter 9, Unit 9.34, (2010).
Perfetto, S.P., Ambrozak, D., Nguyen, R., Chattopadhyay, P.K. & Roederer, M. Quality assurance for polychromatic flow cytometry using a suite of calibration beads. Nat. Protoc. 7, 2067–2079 (2012).
Roederer, M. How many events is enough? Are you positive? Cytometry A 73, 384–385 (2008).
Raj, A. & Tyagi, S. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. Methods Enzymol. 472, 365–386 (2010).
Shah, K. & Tyagi, S. Barriers to transmission of transcriptional noise in a c-fos c-jun pathway. Mol. Syst. Biol. 9, 687 (2013).
Caligiuri, M.A. Human natural killer cells. Blood 112, 461–469 (2008).
Sester, U. et al. Whole-blood flow-cytometric analysis of antigen-specific CD4 T-cell cytokine profiles distinguishes active tuberculosis from non-active states. PLoS One 6, e17813 (2011).
Acknowledgements
This work was supported by grants from the National Institutes of Health (AI-104615, AI-106036 and AI-124691) and by an intramural grant from the Rutgers Office of Research and Economic Development/New Jersey Health Foundation. We are indebted to S.A.E. Marras of the Public Health Research Institute and S. Singh of the Rutgers NJMS Flow Cytometry and Immunology Core Laboratory for constant assistance, advice and superb technical support. We are also grateful to K. Drlica for critical comments on the manuscript.
Author information
Authors and Affiliations
Contributions
R.A., M.L.G. and S.T. conceived and designed the experiments. R.A. and S.T. performed the experiments. R.A., M.L.G. and S.T. analyzed the data. Y.B. and R.P. contributed to the initial FISH-Flow protocol. F.R., K.L. and P.V. contributed to the initial FISH-Flow experiments. D.M. and J.S. developed the modified LWA instrument and contributed to semiautomated protocol design and data analysis. Y.Z. and G.S.Y. designed and provided cells for the concurrent FISH-Flow/ICS time-course experiments. S.T. developed the RNA FISH probes. A.A.L. obtained blood samples from patients. M.L.G. supervised the study. R.A., M.L.G. and S.T. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Rutgers University receives royalties from the sale of prelabeled sm-FISH probes by Biosearch Technologies, which markets them as Stellaris probes. A fraction of these proceeds is distributed to S.T.'s laboratory for research and to him personally. D.M. and J.S. are currently employed by BD Biosciences. Additional patent applications related to this technology coauthored by Y.B., M.L.G., S.T. and R.P. are pending. These proceeds, affiliations and patent applications do not influence the conclusions of this research.
Rights and permissions
About this article
Cite this article
Arrigucci, R., Bushkin, Y., Radford, F. et al. FISH-Flow, a protocol for the concurrent detection of mRNA and protein in single cells using fluorescence in situ hybridization and flow cytometry. Nat Protoc 12, 1245–1260 (2017). https://doi.org/10.1038/nprot.2017.039
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2017.039
This article is cited by
-
Dichotomy between the humoral and cellular responses elicited by mRNA and adenoviral vector vaccines against SARS-CoV-2
BMC Medicine (2022)
-
S100A8-mediated metabolic adaptation controls HIV-1 persistence in macrophages in vivo
Nature Communications (2022)
-
An outlook on fluorescent in situ hybridization coupled to flow cytometry as a versatile technique to evaluate the effects of foods and dietary interventions on gut microbiota
Archives of Microbiology (2022)
-
Microdroplet-based one-step RT-PCR for ultrahigh throughput single-cell multiplex gene expression analysis and rare cell detection
Scientific Reports (2021)
-
Comparison of bias and resolvability in single-cell and single-transcript methods
Communications Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.