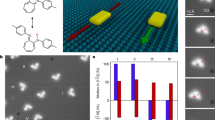
Abstract
Intracellular protein motors have evolved to perform specific tasks critical to the function of cells such as intracellular trafficking and cell division1,2. Kinesin and dynein motors, for example, transport cargoes in living cells by walking along microtubules powered by adenosine triphosphate hydrolysis3,4. These motors can make discrete 8 nm centre-of-mass steps and can travel over 1 µm by changing their conformations during the course of adenosine triphosphate binding, hydrolysis and product release5,6. Inspired by such biological machines, synthetic analogues have been developed including self-assembled DNA walkers that can make stepwise movements on RNA/DNA substrates7,8,9,10,11,12 or can function as programmable assembly lines13. Here, we show that motors based on RNA-cleaving DNA enzymes14 can transport nanoparticle cargoes—CdS nanocrystals in this case—along single-walled carbon nanotubes. Our motors extract chemical energy from RNA molecules decorated on the nanotubes and use that energy to fuel autonomous, processive walking through a series of conformational changes along the one-dimensional track. The walking is controllable and adapts to changes in the local environment, which allows us to remotely direct ‘go’ and ‘stop’ actions. The translocation of individual motors can be visualized in real time using the visible fluorescence of the cargo nanoparticle and the near-infared emission of the carbon-nanotube track. We observed unidirectional movements of the molecular motors over 3 µm with a translocation velocity on the order of 1 nm min−1 under our experimental conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Vale, R. D. The molecular motor toolbox for intracellular transport. Cell 112, 467–480 (2003).
Finer, J. T., Simmons, R. M. & Spudich, J. A. Single myosin molecule mechanics—piconewton forces and nanometre steps. Nature 368, 113–119 (1994).
Hirokawa, N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279, 519–526 (1998).
Howard, J., Hudspeth, A. J. & Vale, R. D. Movement of microtubules by single kinesin molecules. Nature 342, 154–158 (1989).
Block, S. M., Goldstein, L. S. B. & Schnapp, B. J. Bead movement by single kinesin molecules studied with optical tweezers. Nature 348, 348–352 (1990).
Yildiz, A., Tomishige, M., Vale, R. D. & Selvin, P. R. Kinesin walks hand-over-hand. Science 303, 676–678 (2004).
Bath, J., Green, S. J. & Turberfield, A. J. A Free-running DNA motor powered by a nicking enzyme. Angew. Chem. Int. Ed. 44, 4358–4361 (2005).
Yin, P., Choi, H. M. T., Calvert, C. R. & Pierce, N. A. Programming biomolecular self-assembly pathways. Nature 451, 318–322 (2008).
Omabegho, T., Sha, R. & Seeman, N. C. A bipedal DNA Brownian motor with coordinated legs. Science 324, 67–71 (2009).
He, Y. & Liu, D. R. Autonomous multistep organic synthesis in a single isothermal solution mediated by a DNA walker. Nature Nanotech. 5, 778–782 (2010).
Lund, K. et al. Molecular robots guided by prescriptive landscapes. Nature 465, 206–210 (2010).
Wickham, S. F. J. et al. Direct observation of stepwise movement of a synthetic molecular transporter. Nature Nanotech. 6, 166–169 (2011).
Gu, H. Z., Chao, J., Xiao, S. J. & Seeman, N. C. A proximity-based programmable DNA nanoscale assembly line. Nature 465, 202–205 (2010).
Santoro, S. W. & Joyce, G. F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA 94, 4262–4266 (1997).
Santoro, S. W. & Joyce, G. F. Mechanism and utility of an RNA-cleaving DNA enzyme. Biochemistry 37, 13330–13342 (1998).
Tian, Y., He, Y., Chen, Y., Yin, P. & Mao, C. D. Molecular devices—a DNAzyme that walks processively and autonomously along a one-dimensional track. Angew. Chem. Int. Ed. 44, 4355–4358 (2005).
Zhang, D. Y. & Seelig, G. Dynamic DNA nanotechnology using strand-displacement reactions. Nature Chem. 3, 103–113 (2011).
Block, S. M. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys. J. 92, 2986–2995 (2007).
Jeng, E. S., Moll, A. E., Roy, A. C., Gastala, J. B. & Strano, M. S. Detection of DNA hybridization using the near-infrared band-gap fluorescence of single-walled carbon nanotubes. Nano Lett. 6, 371–375 (2006).
Cha, T.-G. et al. Understanding oligonucleotide-templated nanocrystals: growth mechanisms and surface properties. ACS Nano 6, 8136–8143 (2012).
Tsyboulski, D. A., Bachilo, S. M. & Weisman, R. B. Versatile visualization of individual single-walled carbon nanotubes with near-infrared fluorescence microscopy. Nano Lett. 5, 975–979 (2005).
Cognet, L. et al. Stepwise qeunching of exciton fluorescence in carbon nanotubes by single-molecule reactions. Science 316, 1465–1468 (2007).
Dahan, M. et al. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science 302, 442–445 (2003).
Carlson, L. J. & Krauss, T. D. Photophysics of individual single-walled carbon nanotubes. Acc. Chem. Res. 41, 235–243 (2008).
Alivisatos, A. P. The use of nanocrystals in biological detection. Nature Biotechnol. 22, 47–52 (2004).
Choi, J. H. & Strano, M. S. Solvatochromism in single-walled carbon nanotubes. Appl. Phys. Lett. 90, 223114 (2007).
Bachilo, S. M. et al. Structure-assigned optical spectra of single walled carbon nanotubes. Science 298, 2361–2366 (2002).
Schubert, S. et al. RNA cleaving ‘10–23’ DNAzymes with enhanced stability and activity. Nucleic Acids Res. 31, 5982–5992 (2003).
Kou, S. C., Cherayil, B. J., Min, W., English, B. P. & Xie, X. S. Single-molecule Michaelis–Menten equations. J. Phys. Chem. B 109, 19068–19081 (2005).
Panyutin, I. G. & Hsieh, P. The kinetics of spontaneous DNA branch migration. Proc. Natl Acad. Sci. USA 91, 2021–2025 (1994).
Acknowledgements
The authors thank H.N. Robinson and M.C. Akatay for help with synthesis and TEM characterization of CdS nanocrystals. This work was supported by the US Office of Naval Research (awards N00014-11-1-0220 and N00014-12-1-0829). J.H.C. acknowledges a National Science Foundation Career award.
Author information
Authors and Affiliations
Contributions
J.H.C. and C.M. conceived the idea. J.H.C. and T.G.C. designed the research. T.G.C. synthesized materials and performed the motor translocation experiments with assistance from J.P. and J.S. H.C. and J.P. collected AFM images. C.M. and X.L designed the oligonucleotide motifs. T.G.C. and J.H.C. developed the kinetic model and analysed the data. J.H.C. and T.G.C. co-wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary Information (PDF 4212 kb)
Rights and permissions
About this article
Cite this article
Cha, TG., Pan, J., Chen, H. et al. A synthetic DNA motor that transports nanoparticles along carbon nanotubes. Nature Nanotech 9, 39–43 (2014). https://doi.org/10.1038/nnano.2013.257
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2013.257
This article is cited by
-
Enzyme-driven Nanorobots Walking Along Predesigned Tracks on the DNA Origami for Cargo Transport and Catalysis
Chemical Research in Chinese Universities (2024)
-
Construction and Application of DNAzyme-based Nanodevices
Chemical Research in Chinese Universities (2023)
-
A novel binding-induced DNAzyme motor triggered by survivin mRNA
Analytical and Bioanalytical Chemistry (2022)
-
Cellular macromolecules-tethered DNA walking indexing to explore nanoenvironments of chromatin modifications
Nature Communications (2021)
-
Nucleic Acids-based Functional Nanomaterials for Bioimaging
Journal of Analysis and Testing (2021)