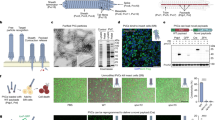
Abstract
Nanoparticles and bacteria can be used, independently, to deliver genes and proteins into mammalian cells for monitoring or altering gene expression and protein production. Here, we show the simultaneous use of nanoparticles and bacteria to deliver DNA-based model drug molecules in vivo and in vitro. In our approach, cargo (in this case, a fluorescent or a bioluminescent gene) is loaded onto the nanoparticles, which are carried on the bacteria surface. When incubated with cells, the cargo-carrying bacteria (‘microbots’) were internalized by the cells, and the genes released from the nanoparticles were expressed in the cells. Mice injected with microbots also successfully expressed the genes as seen by the luminescence in different organs. This new approach may be used to deliver different types of cargo into live animals and a variety of cells in culture without the need for complicated genetic manipulations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Chan, W. C. & Nie, S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science 281, 2016–2018 (1998).
Gao, X. et al. In vivo molecular and cellular imaging with quantum dots. Curr. Opin. Biotech. 16, 63–72 (2005).
Lin, Z., Su, X., Mu, Y. & Jin, Q. Methods for labeling quantum dots to biomolecules. J. Nanosci. Nanotech. 4, 641–645 (2004).
Voura, E. B., Jaiswal, J. K., Mattoussi, H. & Simon, S. M. Tracking metastatic tumor cell extravasation with quantum dot nanocrystals and fluorescence emission-scanning microscopy. Nature Med. 10, 993–998 (2004).
Ballou, B., Ernst, L. A. & Waggoner, A. S. Fluorescence imaging of tumors in vivo. Curr. Med. Chem. 12, 795–805 (2005).
West, J. L. & Halas, N. J. Engineered nanomaterials for biophotonics applications: improving sensing, imaging, and therapeutics. Annu. Rev. Biomed. Eng. 5, 285–292 (2003).
Seppenwoolde, J. H. et al. Internal radiation therapy of liver tumors: qualitative and quantitative magnetic resonance imaging of the biodistribution of holmium-loaded microspheres in animal models. Magn. Reson. Med. 53, 76–84 (2005).
Hattori, Y. & Maitani, Y. Enhanced in vitro DNA transfection efficiency by novel folate-linked nanoparticles in human prostate cancer and oral cancer. J. Control. Release 97, 173–183 (2004).
Jain, R. K. Haemodynamic and transport barriers to the treatment of solid tumors. Int. J. Radiat. Biol. 60, 85–100 (1991).
Maeda, H., Wu, J., Sawa, T., Matsumura, Y. & Hori, K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J. Control. Rel. 65, 271–284 (2000).
Vassaux, G., Nitcheu, J., Jezzard, S. & Lemoine, N. R. Bacterial gene therapy strategies. J. Pathol. 208, 290–298 (2006).
Vazquez-Boland, J. A. et al. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640 (2001).
Finlay, B. B. & Cossart, P. Exploitation of mammalian host cell functions by bacterial pathogens. Science 276, 718–725 (1997).
Vazquez-Boland, J. A., Dominguez-Bernal, G., Gonzalez-Zorn, B., Kreft, J. & Goebel, W. Pathogenicity islands and virulence evolution in Listeria. Microbes Infect. 3, 571–584 (2001).
Hamon, M. L., Bierne, H. & Cossart, P. Listeria monocytogenes: a multifaceted model. Nature Rev. Microbiol. 4, 423–434 (2006).
Pilgrim, S. et al. Bactofection of mammalian cells by Listeria monocytogenes: improvement and mechanism of DNA delivery. Gene Ther. 10, 2036–2045 (2003).
Dietrich, G. et al. Delivery of antigen encoding plasmid DNA into the cytosol of macrophages by attenuated suicide Listeria monocytogenes. Nature Biotechnol. 16, 862–866 (1998).
Sizemore, D. R., Branstrom, A. A. & Sadoff, J. C. Attenuated Shigella as a DNA delivery vehicle for DNA-mediated immunization. Science 270, 299–302 (1995).
Darji, A. et al. Oral somatic transgene vaccination using attenuated S. typhimurium. Cell 91, 765–775 (1997).
Paglia, P., Medina, E., Arioli, I., Guzman, C. A. & Colombo, M. P. Gene transfer in dendritic cells, induced by oral DNA vaccination with Salmonella typhimurium, results in protective immunity against a murine fibrosarcoma. Blood 92, 3172–3176 (1998).
Yu, Y. A. et al. Visualization of tumors and metastases in live animals with bacteria and vaccinia virus encoding light-emitting proteins. Nature Biotechnol. 22, 313–320 (2004).
Bermudes, D., Zheng, L. M. & King, I. C. Live bacteria as anticancer agents and tumor-selective protein delivery vectors. Curr. Opin. Drug Discov. Devel. 5, 194–199 (2002).
Loeffler, D. I., Schoen, C. U., Goebel, W. & Pilgrim, S. Comparison of different live vaccine strategies in vivo for delivery of protein antigen or antigen-encoding DNA and mRNA by virulence-attenuated Listeria monocytogenes. Infect. Immun. 74, 3946–3957 (2006).
Souders, N. C., Verch, T. & Paterson, Y. In vivo bactofection: Listeria can function as a DNA-cancer vaccine. DNA & Cell Biol. 25, 142–151 (2006).
Souders, N. C., Sewell, D. A., Pan, Z. K., Hussain, S. F., Rodriguez, A., Wallecha, A. & Paterson, Y. Listeria-based vaccines can overcome tolerance by expanding low avidity CD8+ T cells capable of eradicating a solid tumor in a transgenic mouse model of cancer. Cancer Immun. 7, 2–12 (2007).
Weber, W. & Fussenegger, M. Inducible gene expression in mammalian cells and mice. Methods Mol. Biol. 267, 451–466 (2004).
Bhunia, A. K et al. Development and characterization of a monoclonal antibody specific for Listeria monocytogenes and Listeria innocua. Infect. Immun. 59, 3176–3184 (1991).
Geng, T. et al. Expression of cellular antigens of Listeria monocytogenes that react with monoclonal antibodies C11E9 and EM-7G1 under acid-, salt- or temperature-induced stress environments. J. Appl. Microbiol. 95, 762–772 (2003).
Geng, T., Hahm, B. K. & Bhunia, A. K. Selective enrichment media affect the antibody-based detection of stress-exposed Listeria monocytogenes due to differential expression of antibody-reactive antigens identified by protein sequencing. J. Food Prot. 69, 1879–1886 (2006).
Drevets, D.A. Dissemination of Listeria monocytogenes by infected phagocytes. Infect. Immun. 67, 3512–3517 (1999).
Bron, P. A., Monk, I. R., Corr, S. C., Hill, C. & Gahan, C. G. Novel luciferase reporter system for in vitro and organ-specific monitoring of differential gene expression in Listeria monocytogenes. Appl. Environ. Microbiol. 72, 2876–2884 (2006).
Hardy, J., Margolis, J. J. & Contag, C. H. Induced biliary excretion of Listeria monocytogenes. Infect Immun. 74, 1819–1827 (2006).
Miller, A. D. The problem with cationic liposome/micelle-based non-viral vector systems for gene therapy. Curr. Med. Chem. 10, 1195–2110 (2003).
El-Aneed, A. An overview of current delivery systems in cancer gene therapy. J. Control. Rel. 94, 1–14 (2004).
Lechardeur, D. & Lukacs, G. L. Intracellular barriers to non-viral gene transfer. Curr. Gene Ther. 2, 183–194 (2002).
Cheong, I. et al. A bacterial protein enhances the release and efficacy of liposomal cancer drugs. Science 314, 1308–1311 (2006).
Michl, P. and Gress, T. M. Bacteria and bacterial toxins as therapeutic agents for solid tumors. Curr. Cancer Drug Targets. 4, 689–702 (2004).
Braun, L., Nato, F., Payrastre, B., Mazie, J. C. & Cossart, P. The 213-amino-acid leucine-rich repeat region of the Listeria monocytogenes InlB protein is sufficient for entry into mammalian cells, stimulation of PI 3-kinase and membrane ruffling. Mol. Microbiol. 34, 10–23 (1999).
Tang, P., Foubister, V., Pucciarelli, G. & Finlay, B. B. Methods to study bacterial invasion. J. Microbiol. Methods 18, 227–240 (1993).
Zreiqat, H., Sungaran, R., Howlett, C. R. & Markovic, B. Quantitative aspects of an in situ hybridization procedure for detecting mRNAs in cells using 96-well microplates. Mol. Biotechnol. 10, 107–113 (1998).
Acknowledgements
The authors would like to thank C. Koons, Drug Discovery Shared Resource of Purdue Cancer Center, for her help with the in vivo studies, S. Leavesly for his inputs in the initial bioluminescence imaging studies, C. Buck for assisting in the use of the facilities at Bindley Biosciences Center, and the Weldon School of Biomedical Engineering for funding the work. D.A. was supported by funds from NIH NIBIB.
Author information
Authors and Affiliations
Contributions
D.A. and R.B. designed the experiments. D.A. performed and was involved in all aspects of the experiments; J.S. performed confocal and fluorescence imaging; K.R. and J.P.R. performed the flow cytometery; and D.S. performed the SEM imaging. D.A., K.B. and A.B. designed and performed the cytotoxicity studies. S.M. assisted in in vivo studies. D.A. and R.B. co-wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary methods, table and figures S1-S7 (PDF 382 kb)
Supplementary Information
Supplementary figure S1a video (AVI 1601 kb)
Supplementary Information
Supplementary figure S1b video (AVI 1601 kb)
Supplementary Information
Supplementary figure S2a video (AVI 2745 kb)
Supplementary Information
Supplementary figure S2b video (AVI 1914 kb)
Supplementary Information
Supplementary figure S3 video (AVI 2133 kb)
Rights and permissions
About this article
Cite this article
Akin, D., Sturgis, J., Ragheb, K. et al. Bacteria-mediated delivery of nanoparticles and cargo into cells. Nature Nanotech 2, 441–449 (2007). https://doi.org/10.1038/nnano.2007.149
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2007.149
This article is cited by
-
Bacterial therapies at the interface of synthetic biology and nanomedicine
Nature Reviews Bioengineering (2023)
-
In vivo bioluminescence imaging of natural bacteria within deep tissues via ATP-binding cassette sugar transporter
Nature Communications (2023)
-
Modulating gut microbiota using nanotechnology to increase anticancer efficacy of the treatments
Macromolecular Research (2023)
-
Remodeling nanodroplets into hierarchical mesoporous silica nanoreactors with multiple chambers
Nature Communications (2022)
-
Spatiotemporal control of engineered bacteria to express interferon-γ by focused ultrasound for tumor immunotherapy
Nature Communications (2022)