Abstract
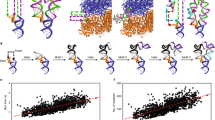
We present fragment assembly of RNA with full-atom refinement (FARFAR), a Rosetta framework for predicting and designing noncanonical motifs that define RNA tertiary structure. In a test set of thirty-two 6–20-nucleotide motifs, FARFAR recapitulated 50% of the experimental structures at near-atomic accuracy. Sequence redesign calculations recovered native bases at 65% of residues engaged in noncanonical interactions, and we experimentally validated mutations predicted to stabilize a signal recognition particle domain.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Gesteland, R.F., Cech, T.R. & Atkins, J.F. The RNA World: The Nature of Modern RNA Suggests a Prebiotic RNA World (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA, 2006).
Shapiro, B.A., Yingling, Y.G., Kasprzak, W. & Bindewald, E. Curr. Opin. Struct. Biol. 17, 157–165 (2007).
Moore, P.B. Annu. Rev. Biochem. 68, 287–300 (1999).
Brion, P. & Westhof, E. Annu. Rev. Biophys. Biomol. Struct. 26, 113–137 (1997).
Das, R. & Baker, D. Proc. Natl. Acad. Sci. USA 104, 14664–14669 (2007).
Ding, F. et al. RNA 14, 1164–1173 (2008).
Massire, C. & Westhof, E. J. Mol. Graph Model. 16, 197–205 (1998).
Sharma, S., Ding, F. & Dokholyan, N.V. Bioinformatics 24, 1951–1952 (2008).
Parisien, M. & Major, F. Nature 452, 51–55 (2008).
Breaker, R.R. Nature 432, 838–845 (2004).
Win, M.N. & Smolke, C.D. Proc. Natl. Acad. Sci. USA 104, 14283–14288 (2007).
Jaeger, L., Westhof, E. & Leontis, N.B. Nucleic Acids Res. 29, 455–463 (2001).
Rohl, C.A., Strauss, C.E., Misura, K.M. & Baker, D. Methods Enzymol. 383, 66–93 (2004).
Klein, D.J., Schmeing, T.M., Moore, P.B. & Steitz, T.A. EMBO J. 20, 4214–4221 (2001).
Leontis, N.B. & Westhof, E. RNA 7, 499–512 (2001).
Boas, F.E. & Harbury, P.B. Curr. Opin. Struct. Biol. 17, 199–204 (2007).
Larsen, N. & Zwieb, C. Nucleic Acids Res. 19, 209–215 (1991).
Crooks, G.E., Hon, G., Chandonia, J.M. & Brenner, S.E. Genome Res. 14, 1188–1190 (2004).
Baeyens, K.J., De Bondt, H.L., Pardi, A. & Holbrook, S.R. Proc. Natl. Acad. Sci. USA 93, 12851–12855 (1996).
Bradley, P. & Baker, D. Proteins 65, 922–929 (2006).
Das, R. & Baker, D. Annu. Rev. Biochem. 77, 363–382 (2008).
Draper, D.E., Grilley, D. & Soto, A.M. Annu. Rev. Biophys. Biomol. Struct. 34, 221–243 (2005).
Qiu, D., Shenkin, P.S., Hollinger, F.P. & Still, W.C. J. Phys. Chem. A 101, 3005–3014 (1997).
Brooks, B.R. et al. J. Comput. Chem. 4, 187–217 (1983).
MacKerell, A.D.J. et al. J. Phys. Chem. B 102, 3586 (1998).
Lee, M.S., Salsbury, F.R.J. & Brooks, C.L.I. J. Chem. Phys. 116, 10606 (2002).
Lee, M., Feig, M., Salsbury, F.J. & Brooks, C.R. J. Comput. Chem. 24, 1348–1356 (2003).
Feig, M., Karanicolas, J. & Brooks, C. J. Mol. Graph. Model. 222, 377–395 (2004).
Yang, H. et al. Nucleic Acids Res. 31, 3450–3460 (2003).
Das, R., Laederach, A., Pearlman, S.M., Herschlag, D. & Altman, R.B. RNA 11, 344–354 (2005).
Acknowledgements
We thank contributors to the current Rosetta codebase, local computer administrators D. Alonso and K. Laidig, the BioX2 cluster (US National Science Foundation award CNS-0619926) and TeraGrid computing resources for enabling rapid development of macromolecular modeling methods; and K. Sjölander for suggesting the acronym FARFAR. This work was supported by the Jane Coffin Childs and Burroughs-Wellcome Foundations (R.D.), the Damon Runyon Cancer Research Foundation (J.K.) and the Howard Hughes Medical Institute (D.B.).
Author information
Authors and Affiliations
Contributions
R.D. designed research, implemented the method, analyzed data and prepared the manuscript; J.K. designed research and implemented the method; and D.B. designed research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9 and Supplementary Tables 1–3 (PDF 6340 kb)
Rights and permissions
About this article
Cite this article
Das, R., Karanicolas, J. & Baker, D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nat Methods 7, 291–294 (2010). https://doi.org/10.1038/nmeth.1433
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1433
This article is cited by
-
Crystal structure of a highly conserved enteroviral 5′ cloverleaf RNA replication element
Nature Communications (2023)
-
trRosettaRNA: automated prediction of RNA 3D structure with transformer network
Nature Communications (2023)
-
RNA structure probing reveals the structural basis of Dicer binding and cleavage
Nature Communications (2021)
-
Pairing a high-resolution statistical potential with a nucleobase-centric sampling algorithm for improving RNA model refinement
Nature Communications (2021)