Abstract
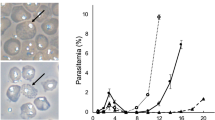
Malaria is a major cause of global morbidity and mortality, and new strategies for treating and preventing this disease are needed. Here we show that the Streptococcus pyogenes Cas9 DNA endonuclease and single guide RNAs (sgRNAs) produced using T7 RNA polymerase (T7 RNAP) efficiently edit the Plasmodium falciparum genome. Targeting the genes encoding native knob-associated histidine-rich protein (kahrp) and erythrocyte binding antigen 175 (eba-175), we achieved high (≥50–100%) gene disruption frequencies within the usual time frame for generating transgenic parasites.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Crabb, B.S. et al. Cell 89, 287–296 (1997).
Wu, Y., Kirkman, L.A. & Wellems, T.E. Proc. Natl. Acad. Sci. USA 93, 1130–1134 (1996).
Straimer, J. et al. Nat. Methods 9, 993–998 (2012).
Maeder, M.L., Thibodeau-Beganny, S., Sander, J.D., Voytas, D.F. & Joung, J.K. Nat. Protoc. 4, 1471–1501 (2009).
Sander, J.D. et al. Nat. Methods 8, 67–69 (2011).
Bassett, A.R., Tibbit, C., Ponting, C.P. & Liu, J.L. Cell Rep. 4, 220–228 (2013).
Cong, L. et al. Science 339, 819–823 (2013).
Dickinson, D.J., Ward, J.D., Reiner, D.J. & Goldstein, B. Nat. Methods 10, 1028–1034 (2013).
Hwang, W.Y. et al. Nat. Biotechnol. 31, 227–229 (2013).
Mali, P. et al. Science 339, 823–826 (2013).
Ghorbal, M. et al. Nat. Biotechnol. 10.1038/nbt.2925 (1 June 2014).
Russell, K., Hasenkamp, S., Emes, R. & Horrocks, P. BMC Genomics 14, 267 (2013).
Wakaguri, H. et al. Nucleic Acids Res. 37, D520–D525 (2009).
Wagner, J.C. et al. Malar. J. 12, 373 (2013).
Reed, M.B. et al. Proc. Natl. Acad. Sci. USA 97, 7509–7514 (2000).
Cho, S.W. et al. Genome Res. 24, 132–141 (2014).
Fu, Y. et al. Nat. Biotechnol. 31, 822–826 (2013).
Pattanayak, V. et al. Nat. Biotechnol. 31, 839–843 (2013).
Aravind, L., Iyer, L.M., Wellems, T.E. & Miller, L.H. Cell 115, 771–785 (2003).
Kirkman, L.A., Lawrence, E.A. & Deitsch, K.W. Nucleic Acids Res. 42, 370–379 (2014).
Deitsch, K., Driskill, C. & Wellems, T. Nucleic Acids Res. 29, 850–853 (2001).
Jinek, M. et al. eLife 2, e00471 (2013).
Adjalley, S.H. et al. Proc. Natl. Acad. Sci. USA 108, E1214–E1223 (2011).
Taylor, D.W. et al. Mol. Biochem. Parasitol. 25, 165–174 (1987).
Rug, M., Prescott, S.W., Fernandez, K.M., Cooke, B.M. & Cowman, A.F. Blood 108, 370–378 (2006).
Hsu, P.D. et al. Nat. Biotechnol. 31, 827–832 (2013).
Acknowledgements
We thank D. Taylor (University of Hawaii) for providing the anti-KAHRP mAb 89 antibody used for western blots and N. Watson (W.M. Keck Microscopy Facility at the Whitehead Institute) for technical assistance with SEM imaging. The following reagent was obtained through the MR4 as part of the BEI Resources Repository, NIAID, US National Institutes of Health (NIH): P. falciparum pfGNr malaria expression vector, MRA-462, deposited by C. Plowe. Quantitative PCR analyses were performed through the Genomics and Imaging Facilities Core of the MIT Center for Environmental and Health Sciences, which is supported by NIEHS Center Grant P30-ES002109. This work was supported by NIGMS Biotechnology Training Grant 5-T32-GM08334 (J.C.W.) and NIEHS Training Grant in Toxicology 5-T32-ES007020 (S.J.G.). This material is based upon work supported by the US National Science Foundation Graduate Research Fellowship under grant no. 1122374 (R.J.P.) and an NIH Director's New Innovator Award (1DP2OD007124) to J.C.N. and is funded by a grant from the Bill and Melinda Gates Foundation through the Grand Challenges Explorations initiative (OPP1069759) to J.C.N.
Author information
Authors and Affiliations
Contributions
J.C.W. generated all plasmid reagents and transgenic parasites, and performed all experimental analyses of these lines. R.J.P. performed in vitro Cas9 cleavage assays and deep sequencing. S.J.G. collected SEM imaging data. J.C.W. and J.C.N. designed experiments and analyzed data. J.C.W. and J.C.N. wrote the manuscript with input from R.J.P. and F.Z. J.C.N. supervised the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 In vitro assay to assess cleavage activity of selected sgRNA-T sequences.
Predicted secondary structures for kahrp- and eba-175- sgRNA-Ts with the additional T7 terminator-derived sequence are shown. Cleavage of target DNA by in vitro transcribed kahrp-, eba-175- and pUC19- (negative control) sgRNA-T was assessed in lysates derived from Cas9-expressing HEK293 cells. The numbers flanking the target DNA region used are the base pair locations in the genic locus, where position +1 corresponds to the A in the ATG start codon.
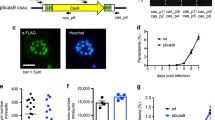
Supplementary Figure 2 Independent experiment to reproduce kahrp locus editing in 3D7 parasites.
(a) RL expression levels were measured over time for no sgRNA-T (negative control) and kahrp-sgRNA-T expressing parasites that were co-transfected with pT7 RNAP-HRkahrp. (b) PCR and Western blot analyses to assess whether kahrp locus editing has occurred in the no sgRNA-T and kahrp-sgRNA-T transfections. SEM images for these parasites are shown in Figure 2f.
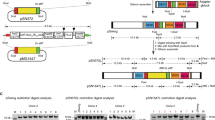
Supplementary Figure 3 CRISPR-Cas9–mediated editing of the P. falciparum eba-175 locus.
(a) Schematic of the plasmids used for genome editing. pCas9-eba175 sgRNA-T and pCas9-pUC19 sgRNA-T encode Cas9 and the bsd selection marker as described in Figure 2a, and produce the eba-175 (test) and pUC19 (control) sgRNA-T respectively, from a T7 promoter-driven cassette. (b) The eba-175 locus and sgRNA-T target site are illustrated, together with the expected editing outcome and location of diagnostic PCR primers to assess locus modification (not drawn to scale). (c) PCR and (d) Southern blot analyses of the pUC19- and eba175- sgRNA-T transfected parasite populations to screen for the expected editing outcome. (e,f) Independent triplicate experiments were analyzed as in c,d above at the population level by PCR (e) and Southern blot (f). DNA isolated from one pUC19-sgRNA-T and two eba175-sgRNA-T transfections (indicated by asterisks) were selected for Southern blot analysis.
Supplementary Figure 4 Assessing the fate of kahrp-sgRNA-T–induced cleavage of the kahrp locus in the absence of a homologous donor plasmid.
Deep sequencing analysis of the region targeted for cleavage by kahrp-sgRNA-T. A representative set (1 of 3 independent experiments) of twenty bacterial clones analyzed by Sanger sequencing is illustrated.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4 and Supplementary Table 1 (PDF 2118 kb)
Rights and permissions
About this article
Cite this article
Wagner, J., Platt, R., Goldfless, S. et al. Efficient CRISPR-Cas9–mediated genome editing in Plasmodium falciparum. Nat Methods 11, 915–918 (2014). https://doi.org/10.1038/nmeth.3063
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.3063
This article is cited by
-
Large DNA fragment knock-in and sequential gene editing in Plasmodium falciparum: a preliminary study using suicide-rescue-based CRISPR/Cas9 system
Molecular and Cellular Biochemistry (2024)
-
Recent Advances in CRISPR/Cas9-Mediated Genome Editing in Leishmania Strains
Acta Parasitologica (2023)
-
Genetic manipulations in helminth parasites
Journal of Parasitic Diseases (2023)
-
CRISPR-Cas9: Taming protozoan parasites with bacterial scissor
Journal of Parasitic Diseases (2022)
-
An integrated platform for genome engineering and gene expression perturbation in Plasmodium falciparum
Scientific Reports (2021)