Abstract
We report an in vitro selection strategy to identify RNA sequences that mediate cap-independent initiation of translation. This method entails mRNA display of trillions of genomic fragments, selection for initiation of translation and high-throughput deep sequencing. We identified >12,000 translation-enhancing elements (TEEs) in the human genome, generated a high-resolution map of human TEE-bearing regions (TBRs), and validated the function of a subset of sequences in vitro and in cultured cells.
Similar content being viewed by others
Main
In eukaryotes, initiation of translation usually follows a cap-dependent mechanism, in which the 43S ribosomal preinitiation complex is recruited to a 7-methylguanosine cap located at the 5′ end of the mRNA strand via recognition of the cap-binding complex eIF4F (refs. 1,2). Although we now have a detailed structural and mechanistic understanding of each step in the cap-dependent process1,2, very little is known about the molecular basis of cap-independent initiation of translation3. Cap-independent translation occurs during normal cellular processes (for example, mitosis and apoptosis) or when the cap-dependent translation machinery is compromised by viral infection or disease4,5. To address this critical gap in our understanding of protein translation, we developed an in vitro selection strategy to identify sequences in the human genome that mediate cap-independent initiation of translation.
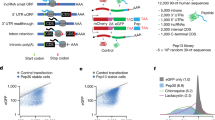
Our selection strategy relies on mRNA display, which is a cell-free method for covalently linking newly translated proteins to their encoding RNA message6. In this approach (Fig. 1a), a genomic library is inserted into the 5′ untranslated region (UTR) of a DNA construct containing the genetic information necessary for mRNA display. The library is in vitro–transcribed to yield a pool of uncapped single-stranded mRNA that is photo-ligated at the 3′ end to a DNA linker containing a 3′ puromycin residue. When translated in vitro, RNA sequences that mediate cap-independent initiation of translation become covalently linked to a peptide affinity tag encoded in the open reading frame. Formation of a chemical bond between newly translated peptides and their encoding mRNA occurs via the natural peptidyl transferase activity of the ribosome, which recognizes puromycin as a tyrosyl-tRNA analog (Fig. 1b). Functional RNAs are then isolated, reverse-transcribed and amplified by PCR to regenerate the pool of DNA for another selection cycle.
(a) A library of human genomic DNA fragments was inserted into a DNA cassette for mRNA display. For each selection round, the dsDNA pool was in vitro–transcribed into single-stranded RNA, conjugated to a DNA-puromycin linker and translated in vitro. Uncapped mRNA sequences that initiate translation of an intact open reading frame become covalently linked to a His-6 protein affinity tag encoded in the RNA message. Functional molecules are recovered, reverse transcribed and amplified by PCR to generate the DNA for the next selection cycle. T7, T7 RNA polymerase promoter; XL, photo–cross-linking site. (b) Schematic of RNA-protein fusion molecule generated via the natural peptidyl transferase activity of the ribosome. (c) Percentage of 35S-labeled fusion molecules recovered from the oligo(dT) and Ni-NTA affinity columns.
We began the selection with a library of ∼1013 RNA-DNA-puromycin molecules containing a random region of genomic fragments (∼150 nucleotides) derived from total human DNA7. We translated the library for 1 h at 30 °C and then incubated the translation mixture overnight at −20 °C under high-salt conditions to promote formation of mRNA-peptide fusions. We isolated the fusions from the crude lysate by oligo(dT) affinity purification, reverse-transcribed the mRNA portion into cDNA to form chimeric cDNA-RNA heteroduplexes and immobilized sequences displaying a His-6 affinity tag on Ni-NTA agarose beads. After washing the beads to remove RNA molecules that did not form mRNA-peptide fusions or did not translate in the correct reading frame, we eluted the remaining mRNA-peptide fusions with imidazole, exchanged the eluate into buffer and performed PCR amplification to reinitiate another selection cycle.
The abundance of mRNA-peptide fusions plateaued after six rounds of mRNA display, indicating that the library had become dominated by sequences that could enhance cap-independent initiation of translation (Fig. 1c). To assess the level of sequence diversity that remained in the pool, we cloned and sequenced individual members from the selection output. We identified 636 unique sequences, 225 of which exhibited 100% identity to the human reference genome (hg18; Supplementary Table 1). The remaining 411 sequences had high homology (85–99% identity) but contained sequence variation that included single nucleotide polymorphisms in addition to small insertions and deletions (Supplementary Table 2). Such variation is expected for individuals in a population, and it is known that functionally relevant sequences can differ between individual genomes8,9.
To test our selected sequences for functional activity in human cells, we modified two luciferase reporter vectors used previously to evaluate translation initiation by adding a promoter sequence specific to our cell-based system10 (Fig. 2a). The first vector contained an unstructured 5′ UTR designed to quantify the activity of TEEs. The second vector contained a stable stem-loop structure (Gibbs free energy (ΔG) = −58 kcal mol−1) upstream of the insert, which blocks translation in the absence of an internal ribosomal entry site (IRES). Translation of both mRNA templates containing a no-insert 13-nucleotide control sequence confirmed that the stem-loop structure inhibited translation (∼99% inhibition) in vitro and in cells (Fig. 2b). Quantitative real-time PCR (qRT-PCR) confirmed that the differences in translation were not caused by differences in RNA expression.
(a) Firefly luciferase reporter (Luc) with or without (+/−) a stable stem-loop structure in the 5′ UTR. p(A)n, polyadenylation signal. (b) Translation efficiency, as measured by luciferase activity, of a no-insert control in the absence and presence of the stem-loop structure, assayed in HeLa cell lysate (in vitro) and in HeLa cells. Error bars, s.d.; n = 3. (c) Translation-enhancing activity of 225 representative sequences after six rounds of in vitro selection, assayed using a luciferase reporter construct in the absence and presence of the stem-loop structure (hairpin) in HeLa cells. Results were compared to data for an unstructured 13-nucleotide insert (red), which defined the basal level of bioluminescence activity for the reporter plasmid. Error bars, s.d.; n = 2. (d) Comparison of 12 high-activity sequences (red) to an equal number of unselected sequences from the starting library (blue) in the absence and presence of the stem-loop structure in HeLa cells and in HeLa cell lysate. Fold enhancement of translation was measured relative to a no insert reporter containing a 13-nucleotide unstructured sequence in place of the TEE. Data shown represents an average of 2 experiments. Raw data are provided in Supplementary Table 3. Luciferase values were normalized to luciferase mRNA data for cell-based experiments in b and d but not in c.
Because cryptic splicing activity is a common cause of IRES misinterpretation11, we used a cytoplasmic expression system that bypasses nuclear expression12. In this system, mammalian cells transfected with an expression vector carrying a vaccinia virus (VACV)-specific promoter are immediately infected with VACV. The virus produces its own RNA polymerase that recognizes the viral promoter and mediates RNA expression in the cytoplasm. We confirmed that nuclear expression did not contribute to translation by measuring the luciferase activity of transfected cells that were not infected with VACV. These cells yielded luciferase values equivalent to those for untreated control cells (data not shown).
Next, we tested perfectly matched sequences for TEE and IRES function in human cells. Using the unstructured vector, we found that the selected sequences produced up to 100-fold more luciferase than the no-insert control (Fig. 2c), demonstrating that our in vitro selection strategy enriched for sequences that enhance translation. Approximately 20% of our TEEs remained functional when tested in the stable stem-loop structure (Fig. 2c), suggesting that a subset of our in vitro–selected TEEs function as IRESs. To ensure that the observed IRES activity was not due to a cryptic promoter13, we screened 20 high-activity sequences in HeLa cells using a vector lacking the VACV promoter. This assay identified 8 sequences with modest to high luciferase activity, indicating that these sequences harbored a cryptic promoter (Supplementary Fig. 1). We considered the remaining 12 sequences to be human IRESs, as their function was not an artifact of RNA splicing or cryptic promoter activity.
We then compared the 12 human IRESs to 12 randomly chosen sequences from the starting library in the unstructured and stem-loop luciferase reporter vectors, both in HeLa cells and in HeLa cell lysates. In the unstructured luciferase reporter system, we observed strong concordance between luciferase assays performed in HeLa cells and in HeLa cell lysates, which resulted in ∼100-fold greater translation-enhancing activity for the 12 human IRESs relative to the randomly chosen sequences from the starting library (Fig. 2d and Supplementary Table 3). We observed a similar trend for the stem-loop luciferase reporter system, which showed that the selected sequences exhibit up to ∼400-fold higher activity in cells and up to ∼100-fold higher activity in vitro than the randomly chosen sequences from the starting library (Fig. 2d and Supplementary Table 3). Collectively, these results establish the ability of our in vitro selection strategy to identify RNA sequences from the human genome that function as efficient translation-enhancing elements, a subset of which function as IRESs.
One caveat of our HeLa cell assay is that the mRNA transcripts likely contain a 5′ cap because of the strong capping enzymes encoded in the VACV genome12. This is not a concern for the hairpin construct as the stem-loop structure blocked cap-dependent initiation of translation (Fig. 2b). However, in the case of the unstructured templates, where a 5′ cap could aid initiation of translation, additional experiments are needed to define the activity of the TEE. We therefore selected 26 sequences that exhibited a range of TEE activities but had no observable IRES activity (Fig. 2c). We then measured their luciferase activity under cap-independent conditions relative to the no-insert control. Consistent with the functional constraints of our in vitro selection, the selected TEEs maintained their activity in the absence of a 5′ cap (Supplementary Fig. 2). In some cases, activity increased considerably when the 5′ cap was missing, suggesting that certain TEEs prefer cap-independent pathways for initiation of translation. This observation provides new insight into the mechanism of initiation of translation where the 5′ cap is thought to inhibit alternate pathways14.
As only a few human TEEs are known15, we performed Illumina deep sequencing on the starting library (round 0, R0) and the selection output (round 6, R6). Sequence analysis revealed that only 2% of the R0 sequences remained in the pool after six rounds of selection. We aligned the R0 and R6 sequences to the reference human genome (hg19) and identified 12,278 unique regions that were enriched by at least tenfold (Online Methods, Supplementary Fig. 3 and Supplementary Table 4). The in vitro–selected TBRs mapped to ∼2 million base pairs. A vast majority of TBRs were shorter than 250 base pairs (99.5%) and were widely dispersed across all 24 chromosomes (Fig. 3a and Supplementary Fig. 4). Of these, 12% (1,532 TBRs) mapped to genomic regions containing known genes, even though genic regions (introns and exons) account for ∼40% of the human genome (Fig. 3b)16. This underabundance in genic regions may be a result of negative selection against TEEs aimed at avoiding disruptive translation in nature, which would be consistent with our results of TEE activity in vitro and in cells (Fig. 2). Moreover, TBRs were preferentially located in 5′ UTRs of genes (threefold over-representation), which would suggest potential functional roles for these elements. We also observed a small but significant enrichment of TBRs in long noncoding RNA regions as compared to the entire human genome (12.2% versus 11.5%, binomial test, P = 0.003), which could lead to the production of novel proteins as these sites are located in intragenic regions of the genome.
(a) Chromosomal ideogram of TBRs with different levels of sequence enrichment between the starting pool (R0) and the selected library (R6): low (10–99-fold), medium (100–999-fold) and high (≥1,000-fold). The blank regions in the chromosome correspond to the unsequenced regions in the reference genome (hg19). Inset, total number of TBRs per chromosome, sorted by enrichment level. FC, fold change. (b) Quantity of TBRs in various genomic regions. TBRs were underrepresented in intragenic and exonic regions (binomial test, both P < 10−16) and overrepresented in 5′ UTRs (binomial test, P < 10−16). CDS, coding sequence. (c) Genomic context of an example TBR residing in an intron of the GRIN2B gene.
Gene Ontology analysis revealed that many TBRs associate with genes involved in signal transduction, cell communication and neurological system development pathways (Supplementary Fig. 5). These functional categories are frequently reported for genes that have undergone adaptive evolution17,18. One example is genes encoding glutamate receptors, which are important for neural communication, memory formation, learning and regulation19. Among the 21 human genes encoding glutamate receptors, eight harbor TBRs in their introns. Of these, two were enriched by more than 1,000-fold after in vitro selection using mRNA display. Some of these sequences are flanked by regions that are highly conserved among species and exhibit transcriptional activity in cells, indicating a possible role for TBRs in the translation of proteins involved in important developmental pathways. One example is a TBR located in an intron of the GRIN2B gene (Fig. 3c). This sequence overlaps with active nucleosome binding sites in the Encyclopedia of DNA Elements (ENCODE) cell lines GM12878 and K562, and is upstream of a highly conserved region among placental mammals. We identified population polymorphisms upstream of, but not within or downstream of, this TBR.
In summary, we present an in vitro selection strategy for searching entire genomes for RNA sequences that enhance cap-independent initiation of translation. Using this technique, we identified >12,000 TEEs in the human genome, generated a high-resolution map of human TEE-bearing regions and validated the function of a subset of sequences in vitro and in cells. Our approach is time-effective, cost-effective, cell line–independent and scalable, making it an effective tool for studying translation mechanisms in other genomes.
Methods
Library assembly and mRNA display selection.
The pool of fragmented human genomic DNA was previously constructed with conserved sequences flanking the random region7. The library was modified by overlap PCR to add all necessary sequence information required for mRNA display. This included a T7 RNA polymerase promoter site upstream of the random region and an open reading frame and photo–cross-linking site downstream of the random region. The open reading frame included a canonical AUG start site followed by a nucleotide sequence encoding a flexible linker and His-6 protein affinity tag. The library was amplified using the forward primer (5′-TTCTAATACGACTCACTATAGGGGGATCCAAGCTTCAGACGTGCCTCACTACG-3′) and reverse primer (5′-ATAGCCGGTGTCCACTTCCATGATGATGGTGATGGTGGGCCATGGCTGAGCTTGACGCTTTGC-3′). For each round of selection, 120 pmol of the dsDNA library was transcribed with T7 RNA polymerase into single-stranded RNA and purified after separation by 10% denaturing urea-PAGE. Purified RNA was photo-ligated to a psoralen-DNA-puromycin linker (5′-psoralen-TAGCCGGTG-(PEG9)2-A15-ACC-puromycin) by irradiating at 366 nm for 15 min. The RNA-DNA-puromycin product was ethanol-precipitated, and the cross-linked RNA (400 pmol) was translated in vitro by incubating the library with micrococcal nuclease–treated rabbit reticulocyte lysate and [35S]methionine for 1 h at 30 °C. The mixture was then incubated overnight at −20 °C in the presence of KCl (600 mM) and MgCl2 (75 mM) to promote formation of fusions. The mRNA-peptide fusion molecules were purified from the crude lysate using oligo (dT)-cellulose beads (NEB) and reverse-transcribed with SuperScript II (Invitrogen) by extending the DNA primer (5′-TTTTTTTTTTTTTTTATCCACTTCCATGATGATGGT-3′) with dNTPs. Fusion molecules containing the correctly translated His-6 tag were isolated on Ni-NTA agarose beads (Qiagen). Functional sequences were recovered by eluting the column with 500 mM imidazole, dialyzing the sample into water and amplifying the cDNA by PCR using previously described overlap PCR primers to add back the necessary sequences for mRNA display. The selection progress was monitored by measuring the fraction of 35S-labeled mRNA-peptide fusions that bound to and eluted from the oligo(dT) and Ni-NTA affinity columns. After six rounds of selection and amplification, the dsDNA library was cloned into a pJET plasmid (Fermentas), and individual isolates were sequenced at the Arizona State University core DNA sequencing facility.
Luciferase reporter plasmids.
A monocistronic luciferase reporter vector with an unstructured 5′ UTR, that contains both a T7 RNA polymerase promoter and a vaccinia virus synthetic late promoter (slp), was constructed from a pT3_R-luc<IRES>F-luc(pA)62 luciferase reporter plasmid10. The vector was first modified using PCR to exchange the T3 promoter with a T7 promoter (forward primer 5′-GATCCCGGGATTAATAACGACTCACTATAGGGAACAAAAGCTGGGTACCGG-3′ and reverse primer 5′-GATCCCGGGTGCGCGCTTGGCGTAATCATGG-3′). The resulting PCR product was cut with SmaI restriction endonuclease and recircularized using T4 DNA ligase. A synthetic dsDNA molecule containing the slp promoter was inserted immediately downstream of the T7 promoter using KpnI and XhoI restriction sites. Finally, the Renilla luciferase gene was removed by PCR using forward primer 5′-ACTAGGATCCGCTTCTGTTGGGAAATGC-3′ and reverse primer 5′-CGCGGATCCAAGCTTATCGATACCGTCGAC-3′. The PCR product was cut with BamHI restriction endonuclease and recircularized using T4 DNA ligase. To assay for IRES activity, two additional luciferase reporter vectors were used, both of which contain a stable stem-loop structure in the 5′ UTR. The first vector was the pT7-stem_F-luc(pA)62 luciferase reporter plasmid described previously2. This plasmid contains a T7 RNA polymerase promoter upstream of the stem-loop. The second vector was constructed by removing the stem-loop structure from pT7-stem_F-luc(pA)62 using StuI and XhoI restriction sites and reciprocally inserting it into the unstructured vector, immediately downstream of the slp promoter. Plasmids to assay for cryptic promoter activity were generated by removing the T7 and slp promoters from the unstructured vector using SmaI and BamHI restriction sites. T4 DNA ligase was then used to insert a 22-nucleotide spacer (5′-ATAGCGCCACCGAGATATCTGG-3′) in place of the promoters. To insert the human genomic sequences into the luciferase reporter vectors, the genomic fragments were amplified by PCR (forward primer 5′-TAGGGGGATCCCAGACGTGCCTCACTACGT-3′ and reverse primer 5′-TGGGCCATGGCTGAGCTTGACGCTTTGCT-3′) to add BamHI and NcoI restriction sites to the 5′ and 3′ ends, respectively. The PCR products were then reciprocally inserted into the vectors immediately upstream of the luciferase coding region by restriction endonuclease digestion.
Cell culture.
HeLa cells, obtained from American Type Culture Collection, were maintained in DMEM (Invitrogen) supplemented with 5% (v/v) FBS (HyClone) and 5 μg/ml gentamicin (Invitrogen). Cells were kept at 37 °C in a humidified atmosphere containing 5% CO2. The cells were free of mycoplasma contamination, as determined by PCR during routine monitoring of cell lysates.
Luciferase reporter assay.
HeLa cells were seeded at a density of 15,000 cells per well in white 96-well plates 18 h before transfection. Cells were transfected with a complex of the luciferase reporter plasmid (200 ng) and Lipofectamine 2000 (0.5 μl) in Opti-MEM (Invitrogen) and immediately infected with the Copenhagen strain (VC-2) of wild-type vaccinia virus at a multiplicity of infection of 5 plaque-forming units per cell. Cells were lysed (6 h after infection) in the 96-well plates, and luciferase activity was measured using the Promega Luciferase Assay System with a Glomax microplate luminometer (Promega). Cell-free characterization of the top translation-enhancing sequences was performed using a Human In Vitro Protein Expression Kit (Pierce). Luciferase expression was achieved following the manufacturer's protocols using 300 ng of linear template for a 2-h transcription at 32 °C followed by a 90-min translation at 30 °C.
RNA characterization.
A portion of the cells used in the luciferase reporter transfection studies were separately lysed to evaluate the quality of the cellular RNA. RNA isolation was performed using the PerfectPure RNA cultured cell kit (5 Prime) according to the manufacturer's protocol. Isolated RNA was reverse-transcribed with an oligo(dT) primer and SuperScript II (Invitrogen). Real-time PCR (iQ SYBR Green Supermix, Bio-Rad) was used to determine the mRNA levels of luciferase (forward primer 5′-GCTGGGCGTTAATCAGAGAG-3′ and reverse primer 5′-GTGTTCGTCTTCGTCCCAGT-3′) as well as the housekeeping gene hypoxanthine-guanine phospho-ribosyltransferase (HPRT, forward primer 5′-TGCTGAGGATTTGGAAAGGGTG-3′ and reverse primer 5′-CCTTGAGCACACAGAGGGCTAC-3′). Using the ΔΔCt method, the amount of luciferase mRNA was normalized to HPRT mRNA levels. Luminescence values were then adjusted according to the normalized luciferase mRNA levels.
Sequence analysis.
An in-house pipeline was used to process Illumina HiSeq sequences. First, base-calling and quality control were performed using the Illumina HiSeq2000 according to the manufacturer's instructions (Supplementary Table 4a). The average length of reads was 80 base pairs (bp). To detect and trim the PCR primers at both ends of each Illumina read, we used the 'cutadapt' program (http://code.google.com/p/cutadapt/) allowing a maximum of two mismatches. Both primers were detected in a vast majority of the reads (85% in R0 and 98% in R6). However, multiple primers were found to be concatenated in some reads, which is common for HiSeq data. For these reads, we used 'cutadapt' iteratively until all primer sequences were trimmed. Finally, reads shorter than 35 bp or longer than 75 bp were discarded because they contained too many or no copies of the primers (Supplementary Table 4b). To ensure correct orientation for all reads, sequences were reverse-complemented if the 5′ primer was present at the 3′ end or the 3′ primer was present at the 5′ end.
All trimmed reads were aligned to the human reference genome build 19 (hg19) using iterative execution of 'bowtie' alignment and end trimmings20. Sequentially, with one base at a time, 16 bp from the 3′ end, 5 bp from the 5′ end and another 15 bp from the 3′ end were trimmed from unaligned reads, which is done to ensure low-quality base calls do not interfere with sequence alignment. In all iterations, 'bowtie' was executed in “-n” mode with “-n 2 -e 70” setting. Reads uniquely mapped to exactly one location, 2–10 locations and more than 10 locations in the hg19 genome were denoted as 'single-copy', 'low-copy' and 'high-copy' reads, respectively (Supplementary Table 4c).
Based on reads mapped to the human genome, we used the command-line version of the CisGenome21 to call peaks where R6 served as the positive sample and R0 served as the negative control sample; parameters were set as “-c 1 -m 10 -w 60 -s 20 -p 0.009948 -br 0 -ssf 0.” Because TEEs are directional, we applied single-strand filtering and labeled a peak as 'forward' or 'reverse' depending on which strand of the genome it resided on. To further reduce spurious peaks, we required a peak to have a strand-specific global false discovery rate less than 10%, total number of reads greater than ten and at least one read present in the R0 library (Supplementary Table 4d). The CisGenome program compared the normalized number of R6 reads with the normalized number of R0 reads in a peak, which represented the fold enrichment level (Supplementary Table 4e). Because repetitive elements can complicate downstream analysis, we focused on peaks derived from single-copy reads. Furthermore, single-copy peaks containing low-complexity sequences were detected using RepeatMasker with parameters “-noint -species human -q.” Peaks with no repeat masked and with more than tenfold enrichment were called putative TBRs (Supplementary Table 4f). Chromosomal distributions of TBRs were converted into ideograms using the Idiographica website22.
We performed bionomical tests for evaluating the null hypothesis that TBRs are randomly distributed in the human genome. In this case, the random probability of a base to belong to a genomic category was first estimated using the RefSeq database to be 0.43, 0.005, 0.005 and 0.57, for genes (all exons and introns), 5′ UTRs, 3′ UTRs and intergenic regions, respectively. We also conducted Gene Ontology enrichment analyses to identify functional categories that were over-represented in the collection of genes found to harbor TBRs (Supplementary Fig. 5). We used Gene Ontology classifications from the PANTHER23 website and applied Bonferroni correction for multiple testing, using a cutoff P value of 10−3. Enriched biological processes were reported (Supplementary Fig. 5). Because the naive library was generated by randomly sampling the genome, longer genes were sampled more often than shorter genes. To account for this gene-length effect, we constructed a background sample from the human genome that matched the length distribution of genes bearing TBRs and redid the Gene Ontology enrichment analysis. This process was repeated ten times. The Bonferroni-corrected P values from each analysis were combined using Fisher's method. Biological processes with P < 0.01 in at least one of these ten gene length–adjusted analyses or with combined P < 0.05 (χ2 test) were highlighted.
Construction and generation of Illumina library.
The Illumina sequencing libraries were generated according to Illumina DNA Sample Kit Instructions (Illumina part 0801-0303). The protocol was modified such that enzymes were obtained from other suppliers, as previously described24. Briefly, DNA from the output of round 6 was end-repaired and phosphorylated using the 'End-It' kit (Epicentre). The blunt, phosphorylated ends were treated with Klenow fragment (3′ to 5′ exo minus; NEB) and dATP to yield a 3′ A overhang for ligation of Illumina's adaptors. After adaptor ligation (LigaFast, Promega) DNA was PCR-amplified with Illumina genomic DNA primers 1.1 and 2.1. The final libraries were isolated (150–300 bp) from an agarose gel to remove residual primers and adaptors. Purified library DNA was captured on an Illumina flow cell for cluster generation and sequenced on an Illumina HiSeq 2000 following the manufacturer's protocols.
References
Sonenberg, N. & Hinnebusch, A.G. Cell 136, 731–745 (2009).
Jackson, R.J., Hellen, C.U.T. & Pestova, T.V. Nat. Rev. Mol. Cell Biol. 10, 113–127 (2010).
Shatsky, I.N., Dmitriev, S.E., Terenin, I.M. & Andreev, D.E. Mol. Cells 30, 285–293 (2010).
Johannes, G., Carter, M.S., Eisen, M.B., Brown, P.O. & Sarnow, P. Proc. Natl. Acad. Sci. USA 96, 13118–13123 (1999).
Spriggs, K.A., Stoneley, M., Bushell, M. & Willis, A.E. Biol. Cell 100, 27–38 (2008).
Roberts, R.W. & Szostak, J.W. Proc. Natl. Acad. Sci. USA 94, 12297–12302 (1997).
Salehi-Ashtiani, K., Luptak, A., Litovchick, A. & Szostak, J.W. Science 313, 1788–1792 (2006).
Korbel, J.O. et al. Science 318, 420–426 (2007).
Kasowski, M. et al. Science 328, 232–235 (2010).
Gilbert, W.V., Zhou, K.H., Butler, T.K. & Doudna, J.A. Science 317, 1224–1227 (2007).
Baranick, B.T. et al. Proc. Natl. Acad. Sci. USA 105, 4733–4738 (2008).
Moss, B. Science 252, 1662–1667 (1991).
Van Eden, M.E., Byrd, M.P., Sherrill, K.W. & Lloyd, R.E. RNA 10, 720–730 (2004).
Mitchell, S.F. et al. Mol. Cell 39, 950–962 (2010).
Mokrejs, M. et al. Nucleic Acids Res. 38, D131–D136 (2010).
Sakharkar, M.K., Chow, V.T. & Kangueane, P. In Silico Biol. 4, 387–393 (2004).
Akey, J.M. Genome Res. 19, 711–722 (2009).
Sabeti, P.C. et al. Science 312, 1614–1620 (2006).
Traynelis, S.F. et al. Pharmacol. Rev. 62, 405–496 (2010).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Genome Biol. 10, R25 (2009).
Ji, H.K. et al. Nat. Biotechnol. 26, 1293–1300 (2008).
Kin, T. & Ono, Y. Methods Biochem. Anal. 23, 2945–2946 (2007).
Thomas, P.D. et al. Genome Res. 13, 2129–2141 (2003).
Auerbach, R.K. et al. Proc. Natl. Acad. Sci. USA 106, 1492–1493 (2009).
Acknowledgements
We thank J. Szostak (Harvard Medical School) and J. Doudna (University of California–Berkeley) for providing the human genome library and luciferase vectors, respectively; N. Pakulis, M. Leon, J. Flores and K. Fenton for technical assistance; G. McInnes for initial bioinformatics analysis; and members of the Chaput laboratory for helpful discussions and comments on the manuscript. This work was supported by US National Institutes of Health to J.C.C. (Eureka Award; GM085530) and S.K. (HG002096-11).
Author information
Authors and Affiliations
Contributions
J.C.C. conceived the project. J.C.C., B.P.W., B.L.J., S.K. and L.L. designed the experiments. B.P.W., B.S., K.W., A.C.L., K.K. and N.B. performed the experiments. J.C.C., B.P.W., A.C.L., K.K., L.L., S.K. and M.S. analyzed the data. J.C.C. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
B.L.J., S.K. and J.C.C. are named inventors on a patent application (PCT/US11/44198) on the technology described in this manuscript.
Supplementary information
Combined PDF
Supplementary Figures 1–5 and Supplementary Tables 1–4 (PDF 2424 kb)
Rights and permissions
About this article
Cite this article
Wellensiek, B., Larsen, A., Stephens, B. et al. Genome-wide profiling of human cap-independent translation-enhancing elements. Nat Methods 10, 747–750 (2013). https://doi.org/10.1038/nmeth.2522
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.2522
This article is cited by
-
Tailor made: the art of therapeutic mRNA design
Nature Reviews Drug Discovery (2024)
-
A researcher’s guide to the galaxy of IRESs
Cellular and Molecular Life Sciences (2017)
-
IRES-dependent translated genes in fungi: computational prediction, phylogenetic conservation and functional association
BMC Genomics (2015)