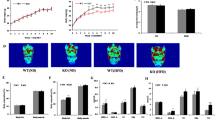
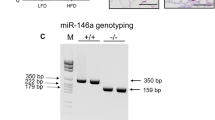
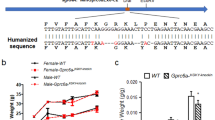
Abstract
Over 40% of microRNAs (miRNAs) are located in introns of protein-coding genes, and many of these intronic miRNAs are co-regulated with their host genes1,2. In such cases of co-regulation, the products of host genes and their intronic miRNAs can cooperate to coordinately regulate biologically important pathways3,4. Therefore, we screened intronic miRNAs dysregulated in the livers of mouse models of obesity to identify previously uncharacterized protein-coding host genes that may contribute to the pathogenesis of obesity-associated insulin resistance and type 2 diabetes mellitus. Our approach revealed that expression of both the gene encoding ectodysplasin A (Eda), the causal gene in X-linked hypohidrotic ectodermal dysplasia (XLHED)5, and its intronic miRNA, miR-676, was increased in the livers of obese mice. Moreover, hepatic EDA expression is increased in obese human subjects and reduced upon weight loss, and its hepatic expression correlates with systemic insulin resistance. We also found that reducing miR-676 expression in db/db mice increases the expression of proteins involved in fatty acid oxidation and reduces the expression of inflammatory signaling components in the liver. Further, we found that Eda expression in mouse liver is controlled via PPARγ and RXR-α, increases in circulation under conditions of obesity, and promotes JNK activation and inhibitory serine phosphorylation of IRS1 in skeletal muscle. In accordance with these findings, gain- and loss-of-function approaches reveal that liver-derived EDA regulates systemic glucose metabolism, suggesting that EDA is a hepatokine that can contribute to impaired skeletal muscle insulin sensitivity in obesity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
BioProject
Gene Expression Omnibus
Referenced accessions
Gene Expression Omnibus
References
Davis, B.N. & Hata, A. Regulation of microRNA biogenesis: a miRiad of mechanisms. Cell Commun. Signal. 7, 18 (2009).
Gromak, N. Intronic microRNAs: a crossroad in gene regulation. Biochem. Soc. Trans. 40, 759–761 (2012).
Najafi-Shoushtari, S.H. et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328, 1566–1569 (2010).
Ma, N. et al. Coexpression of an intronic microRNA and its host gene reveals a potential role for miR-483-5p as an IGF2 partner. Mol. Cell. Endocrinol. 333, 96–101 (2011).
Mikkola, M.L. Molecular aspects of hypohidrotic ectodermal dysplasia. Am. J. Med. Genet. A. 149A, 2031–2036 (2009).
Kornfeld, J.W. et al. Obesity-induced overexpression of miR-802 impairs glucose metabolism through silencing of Hnf1b. Nature 494, 111–115 (2013).
Mikkola, M.L. et al. Ectodysplasin, a protein required for epithelial morphogenesis, is a novel TNF homologue and promotes cell-matrix adhesion. Mech. Dev. 88, 133–146 (1999).
Schneider, P. et al. Mutations leading to X-linked hypohidrotic ectodermal dysplasia affect three major functional domains in the tumor necrosis factor family member ectodysplasin-A. J. Biol. Chem. 276, 18819–18827 (2001).
Hotamisligil, G.S., Shargill, N.S. & Spiegelman, B.M. Adipose expression of tumor necrosis factor-α: direct role in obesity-linked insulin resistance. Science 259, 87–91 (1993).
Srivastava, A.K. et al. The Tabby phenotype is caused by mutation in a mouse homologue of the EDA gene that reveals novel mouse and human exons and encodes a protein (ectodysplasin-A) with collagenous domains. Proc. Natl. Acad. Sci. USA 94, 13069–13074 (1997).
Podzus, J. et al. Ectodysplasin A in biological fluids and diagnosis of ectodermal dysplasia. J. Dent. Res. 96, 217–224 (2017).
Messeguer, X. et al. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333–334 (2002).
Farré, D. et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 31, 3651–3653 (2003).
Hauser, S. et al. Degradation of the peroxisome proliferator-activated receptor γ is linked to ligand-dependent activation. J. Biol. Chem. 275, 18527–18533 (2000).
Vidal-Puig, A. et al. Regulation of PPARγ gene expression by nutrition and obesity in rodents. J. Clin. Invest. 97, 2553–2561 (1996).
Pettinelli, P. & Videla, L.A. Up-regulation of PPAR-γ mRNA expression in the liver of obese patients: an additional reinforcing lipogenic mechanism to SREBP-1c induction. J. Clin. Endocrinol. Metab. 96, 1424–1430 (2011).
Baek, D. et al. The impact of microRNAs on protein output. Nature 455, 64–71 (2008).
Selbach, M. et al. Widespread changes in protein synthesis induced by microRNAs. Nature 455, 58–63 (2008).
Cox, J. & Mann, M. 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13 Suppl 16, S12 (2012).
Huang, W., Sherman, B.T. & Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Hashimoto, T., Cui, C.Y. & Schlessinger, D. Repertoire of mouse ectodysplasin-A (EDA-A) isoforms. Gene 371, 42–51 (2006).
Yan, M. et al. Two–amino acid molecular switch in an epithelial morphogen that regulates binding to two distinct receptors. Science 290, 523–527 (2000).
Lindfors, P.H., Voutilainen, M. & Mikkola, M.L. Ectodysplasin/NF-κB signaling in embryonic mammary gland development. J. Mammary Gland Biol. Neoplasia 18, 165–169 (2013).
Copps, K.D. & White, M.F. Regulation of insulin sensitivity by serine/threonine phosphorylation of insulin receptor substrate proteins IRS1 and IRS2. Diabetologia 55, 2565–2582 (2012).
Newton, K., French, D.M., Yan, M., Frantz, G.D. & Dixit, V.M. Myodegeneration in EDA-A2 transgenic mice is prevented by XEDAR deficiency. Mol. Cell. Biol. 24, 1608–1613 (2004).
Hirosumi, J. et al. A central role for JNK in obesity and insulin resistance. Nature 420, 333–336 (2002).
Sabio, G. et al. Role of muscle c-Jun NH2-terminal kinase 1 in obesity-induced insulin resistance. Mol. Cell. Biol. 30, 106–115 (2010).
Pal, M. et al. Alteration of JNK-1 signaling in skeletal muscle fails to affect glucose homeostasis and obesity-associated insulin resistance in mice. PLoS One 8, e54247 (2013).
Grimm, D. et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 82, 5887–5911 (2008).
Grimm, D. et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441, 537–541 (2006).
Izumiya, Y. et al. Fast/glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell Metab. 7, 159–172 (2008).
Tanikawa, C., Ri, C., Kumar, V., Nakamura, Y. & Matsuda, K. Crosstalk of EDA-A2/XEDAR in the p53 signaling pathway. Mol. Cancer Res. 8, 855–863 (2010).
Kowalczyk-Quintas, C. et al. Pharmacological stimulation of Edar signaling in the adult enhances sebaceous gland size and function. J. Invest. Dermatol. 135, 359–368 (2015).
Cui, C.Y. et al. Inducible mEDA-A1 transgene mediates sebaceous gland hyperplasia and differential formation of two types of mouse hair follicles. Hum. Mol. Genet. 12, 2931–2940 (2003).
Pochi, P.E., Downing, D.T. & Strauss, J.S. Sebaceous gland response in man to prolonged total caloric deprivation. J. Invest. Dermatol. 55, 303–309 (1970).
Chen, H.C., Smith, S.J., Tow, B., Elias, P.M. & Farese, R.V. Jr. Leptin modulates the effects of acyl CoA:diacylglycerol acyltransferase deficiency on murine fur and sebaceous glands. J. Clin. Invest. 109, 175–181 (2002).
Yosipovitch, G., DeVore, A. & Dawn, A. Obesity and the skin: skin physiology and skin manifestations of obesity. J. Am. Acad. Dermatol. 56, 901–916, quiz 917–920 (2007).
Fete, M., Hermann, J., Behrens, J. & Huttner, K.M. X-linked hypohidrotic ectodermal dysplasia (XLHED): clinical and diagnostic insights from an international patient registry. Am. J. Med. Genet. A. 164A, 2437–2442 (2014).
Motil, K.J. et al. Growth characteristics of children with ectodermal dysplasia syndromes. Pediatrics 116, e229–e234 (2005).
Awazawa, M. et al. Adiponectin suppresses hepatic SREBP1c expression in an AdipoR1/LKB1/AMPK dependent pathway. Biochem. Biophys. Res. Commun. 382, 51–56 (2009).
Amrutkar, M. et al. Protein kinase STK25 controls lipid partitioning in hepatocytes and correlates with liver fat content in humans. Diabetologia 59, 341–353 (2016).
Kannt, A. et al. Association of nicotinamide-N-methyltransferase mRNA expression in human adipose tissue and the plasma concentration of its product, 1-methylnicotinamide, with insulin resistance. Diabetologia 58, 799–808 (2015).
Krüger, M. et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell 134, 353–364 (2008).
Wai, T. et al. The membrane scaffold SLP2 anchors a proteolytic hub in mitochondria containing PARL and the i-AAA protease YME1L. EMBO Rep. 17, 1844–1856 (2016).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Cox, J. et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 (2011).
Tyanova, S. et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740 (2016).
Huang, W., Sherman, B.T. & Lempicki, R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Könner, A.C. et al. Insulin action in AgRP-expressing neurons is required for suppression of hepatic glucose production. Cell Metab. 5, 438–449 (2007).
Fisher, S.J. & Kahn, C.R. Insulin signaling is required for insulin's direct and indirect action on hepatic glucose production. J. Clin. Invest. 111, 463–468 (2003).
Ferré, P., Leturque, A., Burnol, A.F., Penicaud, L. & Girard, J. A method to quantify glucose utilization in vivo in skeletal muscle and white adipose tissue of the anaesthetized rat. Biochem. J. 228, 103–110 (1985).
Wagle, P., Nikolic´, M. & Frommolt, P. QuickNGS elevates next-generation sequencing data analysis to a new level of automation. BMC Genomics 16, 487 (2015).
Vizcaíno, J.A. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 11033 (2016).
Acknowledgements
We acknowledge J. Alber, B. Hampel, P. Scholl, N. Spenrath and D. Kutyniok for outstanding technical assistance. We greatly appreciate P. Schneider (Lausanne University) for providing Tabby mice and EDA expression vectors as well as outstanding support in performance of the EDA AlphaLISA assay. We acknowledge J. Wilson (University of Pennsylvania) for providing the pXR8 plasmid and J. Samulski (University of North Carolina) for providing the pXX6-80 plasmid. We acknowledge P. Frommolt for bioinformatics support and H. Büning for support of AAV production. This work was supported by a grant from the German Research Foundation (DFG) (BR 1492/7-1) to J.C.B., and we received funding from DFG within the framework of TRR134 and within the Excellence Initiative by German Federal and State Governments (CECAD). This work was funded (in part) by the Helmholtz Alliance (Imaging and Curing Environmental Metabolic Diseases, ICEMED) through the Initiative and Networking Fund of the Helmholtz Association. J.W.K. greatly appreciates funding from the Emmy Noether program of DFG (KO 4728/1-1). M.A. gratefully acknowledges support from a Manpei Suzuki Diabetes Foundation fellowship.
Author information
Authors and Affiliations
Contributions
M.A. and J.C.B. designed the study and wrote the manuscript. M.A. and P.G. performed experiments, and M.A. analyzed data. E.T. performed the surgeries and with P.G. conducted the hyperinsulinemic–euglycemic clamp experiment. C.B. also supported the hyperinsulinemic–euglycemic clamp studies. H.N. and M.K. performed SILAC and proteomic analyses. P.J.A. contributed to AAV generation. J.S. provided critical reagents. J.A. performed miRNA sequencing, and S.M. analyzed the data. F.T.W. provided JNKSM-C mice. J.-W.K. provided the microarray data. M.B. provided the human samples and clinical data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–4 and Supplementary Figures 1–11 (PDF 28394 kb)
Rights and permissions
About this article
Cite this article
Awazawa, M., Gabel, P., Tsaousidou, E. et al. A microRNA screen reveals that elevated hepatic ectodysplasin A expression contributes to obesity-induced insulin resistance in skeletal muscle. Nat Med 23, 1466–1473 (2017). https://doi.org/10.1038/nm.4420
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4420
This article is cited by
-
EDA2R–NIK signalling promotes muscle atrophy linked to cancer cachexia
Nature (2023)
-
Investigation of candidate genes and mechanisms underlying obesity associated type 2 diabetes mellitus using bioinformatics analysis and screening of small drug molecules
BMC Endocrine Disorders (2021)
-
Diabetes risk assessment with imaging: a radiomics study of abdominal CT
European Radiology (2019)
-
Neuronal activity regulates DROSHA via autophagy in spinal muscular atrophy
Scientific Reports (2018)
-
LncRNAs and miRs as epigenetic signatures in diabetic cardiac fibrosis: new advances and perspectives
Endocrine (2018)