Abstract
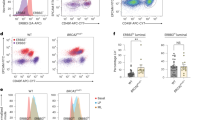
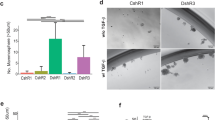
Individuals who have mutations in the breast-cancer-susceptibility gene BRCA1 (hereafter referred to as BRCA1-mutation carriers) frequently undergo prophylactic mastectomy to minimize their risk of breast cancer. The identification of an effective prevention therapy therefore remains a 'holy grail' for the field. Precancerous BRCA1mut/+ tissue harbors an aberrant population of luminal progenitor cells1, and deregulated progesterone signaling has been implicated in BRCA1-associated oncogenesis2,3,4,5. Coupled with the findings that tumor necrosis factor superfamily member 11 (TNFSF11; also known as RANKL) is a key paracrine effector of progesterone signaling6,7,8,9,10 and that RANKL and its receptor TNFRSF11A (also known as RANK) contribute to mammary tumorigenesis11,12,13, we investigated a role for this pathway in the pre-neoplastic phase of BRCA1-mutation carriers. We identified two subsets of luminal progenitors (RANK+ and RANK−) in histologically normal tissue of BRCA1-mutation carriers and showed that RANK+ cells are highly proliferative, have grossly aberrant DNA repair and bear a molecular signature similar to that of basal-like breast cancer. These data suggest that RANK+ and not RANK− progenitors are a key target population in these women. Inhibition of RANKL signaling by treatment with denosumab in three-dimensional breast organoids derived from pre-neoplastic BRCA1mut/+ tissue attenuated progesterone-induced proliferation. Notably, proliferation was markedly reduced in breast biopsies from BRCA1-mutation carriers who were treated with denosumab. Furthermore, inhibition of RANKL in a Brca1-deficient mouse model substantially curtailed mammary tumorigenesis. Taken together, these findings identify a targetable pathway in a putative cell-of-origin population in BRCA1-mutation carriers and implicate RANKL blockade as a promising strategy in the prevention of breast cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Lim, E. et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1-mutation carriers. Nat. Med. 15, 907–913 (2009).
King, T.A. et al. Increased progesterone receptor expression in benign epithelium of BRCA1-related breast cancers. Cancer Res. 64, 5051–5053 (2004).
Ma, Y. et al. The breast cancer susceptibility gene BRCA1 regulates progesterone receptor signaling in mammary epithelial cells. Mol. Endocrinol. 20, 14–34 (2006).
Poole, A.J. et al. Prevention of Brca1-mediated mammary tumorigenesis in mice by a progesterone antagonist. Science 314, 1467–1470 (2006).
Widschwendter, M. et al. The sex hormone system in carriers of BRCA1/2 mutations: a case-control study. Lancet Oncol. 14, 1226–1232 (2013).
Asselin-Labat, M.L. et al. Control of mammary stem cell function by steroid hormone signaling. Nature 465, 798–802 (2010).
Beleut, M. et al. Two distinct mechanisms underlie progesterone-induced proliferation in the mammary gland. Proc. Natl. Acad. Sci. USA 107, 2989–2994 (2010).
Fernandez-Valdivia, R. & Lydon, J.P. From the ranks of mammary progesterone mediators, RANKL takes the spotlight. Mol. Cell. Endocrinol. 357, 91–100 (2012).
Joshi, P.A. et al. Progesterone induces adult mammary stem cell expansion. Nature 465, 803–807 (2010).
Tanos, T. et al. Progesterone–RANKL is a major regulatory axis in the human breast. Sci. Transl. Med. 5, 182ra55 (2013).
Gonzalez-Suarez, E. et al. RANK ligand mediates progestin-induced mammary epithelial proliferation and carcinogenesis. Nature 468, 103–107 (2010).
Schramek, D. et al. Osteoclast differentiation factor RANKL controls development of progestin-driven mammary cancer. Nature 468, 98–102 (2010).
Pellegrini, P. et al. Constitutive activation of RANK disrupts mammary cell fate leading to tumorigenesis. Stem Cells 31, 1954–1965 (2013).
Antoniou, A. et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am. J. Hum. Genet. 72, 1117–1130 (2003).
Perou, C.M. et al. Molecular portraits of human breast tumors. Nature 406, 747–752 (2000).
Turner, N., Tutt, A. & Ashworth, A. Hallmarks of 'BRCAness' in sporadic cancers. Nat. Rev. Cancer 4, 814–819 (2004).
Venkitaraman, A.R. Cancer suppression by the chromosome custodians, BRCA1 and BRCA2. Science 343, 1470–1475 (2014).
Narod, S.A. & Foulkes, W.D. BRCA1 and BRCA2: 1994 and beyond. Nat. Rev. Cancer 4, 665–676 (2004).
Molyneux, G. et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell 7, 403–417 (2010).
Proia, T.A. et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell 8, 149–163 (2011).
Joshi, P.A. et al. RANK signaling amplifies WNT-responsive mammary progenitors through R-SPONDIN1. Stem Cell Rep. 5, 31–44 (2015).
Pal, B. et al. Global changes in the mammary epigenome are induced by hormonal cues and coordinated by Ezh2. Cell Rep. 3, 411–426 (2013).
Eirew, P. et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat. Med. 14, 1384–1389 (2008).
Eirew, P. et al. Aldehyde dehydrogenase activity is a biomarker of primitive normal human mammary luminal cells. Stem Cells 30, 344–348 (2012).
Liu, S. et al. BRCA1 regulates human mammary stem–progenitor cell fate. Proc. Natl. Acad. Sci. USA 105, 1680–1685 (2008).
Wood, C.E. et al. Progestin effects on cell proliferation pathways in the postmenopausal mammary gland. Breast Cancer Res. 15, R62 (2013).
Pathania, S. et al. BRCA1 haploinsufficiency for replication-stress suppression in primary cells. Nat. Commun. 5, 5496 (2014).
Sedic, M. et al. Haploinsufficiency for BRCA1 leads to cell-type-specific genomic instability and premature senescence. Nat. Commun. 6, 7505 (2015).
Kostenuik, P.J. et al. Denosumab, a fully human monoclonal antibody to RANKL, inhibits bone resorption and increases BMD in knock-in mice that express chimeric (murine–human) RANKL. J. Bone Miner. Res. 24, 182–195 (2009).
Shackleton, M. et al. Generation of a functional mammary gland from a single stem cell. Nature 439, 84–88 (2006).
Shehata, M. et al. Phenotypic and functional characterization of the luminal cell hierarchy of the mammary gland. Breast Cancer Res. 14, R134 (2012).
Liu, X. et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc. Natl. Acad. Sci. USA 104, 12111–12116 (2007).
Fu, N.Y. et al. EGF-mediated induction of Mcl-1 at the switch to lactation is essential for alveolar cell survival. Nat. Cell Biol. 17, 365–375 (2015).
Hartmann, L.C. & Lindor, N.M. The role of risk-reducing surgery in hereditary breast and ovarian cancer. N. Engl. J. Med. 374, 454–468 (2016).
Phillips, K.A. & Lindeman, G.J. Breast cancer prevention for BRCA1- and BRCA-mutation carriers: is there a role for tamoxifen? Future Oncol. 10, 499–502 (2014).
Domchek, S.M. et al. Association of risk-reducing surgery in BRCA1- or BRCA2-mutation carriers with cancer risk and mortality. J. Am. Med. Assoc. 304, 967–975 (2010).
Rebbeck, T.R. et al. Prophylactic oophorectomy in carriers of BRCA1 or BRCA2 mutations. N. Engl. J. Med. 346, 1616–1622 (2002).
Heemskerk-Gerritsen, B.A. et al. Breast cancer risk after salpingo-oophorectomy in healthy BRCA1/2-mutation carriers: revisiting the evidence for risk reduction. J. Natl. Cancer Inst. 107, djv033 (2015).
To, C. et al. The PARP inhibitors veliparib and olaparib are effective chemopreventive agents for delaying mammary tumor development in BRCA1-deficient mice. Cancer Prev. Res. (Phila.) 7, 698–707 (2014).
Gnant, M. et al. Adjuvant denosumab in breast cancer (ABCSG-18): a multicenter, randomized, double-blind, placebo-controlled trial. Lancet 386, 433–443 (2015).
Mann, G.J. et al. Analysis of cancer risk and BRCA1 and BRCA2 mutation prevalence in the kConFab familial breast cancer resource. Breast Cancer Res. 8, R12 (2006).
Branstetter, D.G. et al. Denosumab induces tumor reduction and bone formation in patients with giant-cell tumor of bone. Clin. Cancer Res. 18, 4415–4424 (2012).
Wagner, K.U. et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 25, 4323–4330 (1997).
Xu, X. et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumor formation. Nat. Genet. 22, 37–43 (1999).
Jacks, T. et al. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 4, 1–7 (1994).
Oakes, S.R. et al. Sensitization of BCL-2-expressing breast tumors to chemotherapy by the BH3 mimetic ABT-737. Proc. Natl. Acad. Sci. USA 109, 2766–2771 (2012).
Liao, Y., Smyth, G.K. & Shi, W. The Subread aligner: fast, accurate and scalable read-mapping by seed-and-vote. Nucleic Acids Res. 41, e108 (2013).
Liao, Y., Smyth, G.K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M.D. & Oshlack, A. A scaling-normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010).
Ritchie, M.E. et al. limma powers differential expression analyses for RNA sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Law, C.W., Chen, Y., Shi, W. & Smyth, G.K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Smyth, G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, e3 (2004).
Robinson, M.D., McCarthy, D.J. & Smyth, G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Wu, D. et al. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 26, 2176–2182 (2010).
Acknowledgements
We are grateful to the women who generously donated breast tissue for our studies and to the surgical, pathology and tissue bank colleagues for their substantial assistance and support. We thank L. Taylor, K. Shackleton, S. Nightingale and H. Liu for expert assistance; the Animal, FACS, Imaging and Histology facilities at WEHI; C. Perou and K. Hoadley for kind provision of the PAM50 subtypes; and A.Y. Rhee (Amgen Inc.) for assistance with the manuscript submission. We are grateful to H. Thorne and all kConFab staff, members and families for their contributions to this resource and to the Victorian Cancer Biobank (supported by the Victorian Government), which also provided coded breast tissue and data. We thank H. Yasuda for advice on the antibody to mouse RANKL and K.U. Wagner (Eppley Institute, Nebraska) for MMTV-Cre A mice. This work was supported by the Australian National Health and Medical Research Council (NHMRC) (grant numbers 1016701 (J.E.V. and G.J.L.), 1040978 (G.B.M., J.E.V. and G.J.L.) and 1054618 (G.K.S.)), NHMRC Independent Research Institute Infrastructure Support Scheme (IRIISS) (to WEHI; E.N., F.V., B.P., G.G., L.W., S.W.L., G.K.S., J.E.V. and G.J.L.), the Victorian State Government through the Victorian Cancer Agency and Operational Infrastructure Support, the National Breast Cancer Foundation (Australia) (grant numbers NC-13-32 and PS-15-042; to J.E.V. and G.J.L.); Amgen Inc. (J.E.V. and G.J.L.), the Qualtrough Cancer Research Fund (J.E.V. and G.J.L.), the Joan Marshall Breast Cancer Research Fund (J.E.V. and G.J.L.) and the Australian Cancer Research Foundation (to WEHI; E.N., F.V., B.P., G.G., L.W., S.W.L., G.K.S., J.E.V. and G.J.L.). E.N. is supported by a Cancer Council Victoria Scholarship and a Cancer Therapeutics CRC Top-Up Scholarship; B.P. is supported by a VCA Early Career Seed Grant 13035; G.K.S. and G.J.L. are supported by NHMRC Fellowships 1058892 (G.K.S.), 637307 (G.J.L.) and 1078730 (G.J.L.); and J.E.V. is supported by NHMRC Australia Fellowship 1037230.
Author information
Authors and Affiliations
Consortia
Contributions
E.N. designed and performed experiments, carried out data analysis and contributed to manuscript writing; B.P. performed RNA-seq and mRNA studies; D.B. performed pathology scoring and analysis of tumor and normal tissue samples; F.V. performed in vivo studies and analysis; G.G. and G.K.S. carried out bioinformatics analysis; L.W. developed a new scoring assay; and G.J.L. and S.W.L. developed and implemented the investigator-sponsored (ISS) BRCA-D study protocol. No Amgen authors were directly or indirectly involved in the ISS design, implementation, analysis or ISS-derived reported results (Fig. 3d and Supplementary Fig. 4b); G.B.M. provided material and discussions; kConFab provided annotated samples; K.R., L.-Y.H. and R.S. assisted with histological analysis of tumor and normal tissue samples; and J.E.V. and G.J.L. conceived the study, designed experiments and wrote the paper in collaboration with W.C.D.
Corresponding authors
Ethics declarations
Competing interests
D.B., K.R., L.-Y.H., R.S. and W.C.D. were former Amgen employees. D.B., K.R. and L.-Y.H. currently hold Amgen stock. Amgen Inc. contributed reagents and some financial support for this study, including financial support for the investigator-sponsored study, BRCA-D.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1–5 (PDF 7177 kb)
Rights and permissions
About this article
Cite this article
Nolan, E., Vaillant, F., Branstetter, D. et al. RANK ligand as a potential target for breast cancer prevention in BRCA1-mutation carriers. Nat Med 22, 933–939 (2016). https://doi.org/10.1038/nm.4118
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nm.4118
This article is cited by
-
Cell origin of BRCA2-mutant breast cancer
Nature Cell Biology (2024)
-
Identification of aberrant luminal progenitors and mTORC1 as a potential breast cancer prevention target in BRCA2 mutation carriers
Nature Cell Biology (2024)
-
Choices for cancer prevention for women with a BRCA1 mutation? a personal view
Hereditary Cancer in Clinical Practice (2023)
-
Immune cells are increased in normal breast tissues of BRCA1/2 mutation carriers
Breast Cancer Research and Treatment (2023)
-
Differences between zoledronic acid and denosumab for breast cancer treatment
Journal of Bone and Mineral Metabolism (2023)