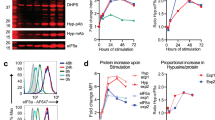
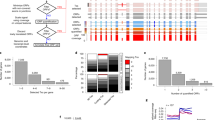
Abstract
Selective translational control of gene expression is emerging as a principal mechanism for the regulation of protein abundance that determines a variety of functions in both the adaptive immune system and the innate immune system. The translation-initiation factor eIF4E acts as a node for such regulation, but non-eIF4E mechanisms are also prevalent. Studies of 'translatomes' (genome-wide pools of translated mRNA) have facilitated mechanistic discoveries by identifying key regulatory components, including transcription factors, that are under translational control. Here we review the current knowledge on mechanisms that regulate translation and thereby modulate immunological function. We further describe approaches for measuring and analyzing translatomes and how such powerful tools can facilitate future insights on the role of translational control in the immune system.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Bava, F.A. et al. CPEB1 coordinates alternative 3′-UTR formation with translational regulation. Nature 495, 121–125 (2013).
Rousseau, D., Kaspar, R., Rosenwald, I., Gehrke, L. & Sonenberg, N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. USA 93, 1065–1070 (1996).
Lindstein, T., June, C.H., Ledbetter, J.A., Stella, G. & Thompson, C.B. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science 244, 339–343 (1989).
Schena, M. et al. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93, 10614–10619 (1996).
Schena, M., Shalon, D., Davis, R.W. & Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470 (1995).
Lockhart, D.J. et al. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14, 1675–1680 (1996).
Larsson, O., Tian, B. & Sonenberg, N. Toward a genome-wide landscape of translational control. Cold Spring Harb. Perspect. Biol. 5, a012302 (2013).
Vogel, C. & Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232 (2012).
Schwanhäusser, B. et al. Global quantification of mammalian gene expression control. Nature 473, 337–342 (2011).
Kristensen, A.R., Gsponer, J. & Foster, L.J. Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol. Syst. Biol. 10.1038/msb.2013.47 (17 September 2013).This study identifies translational control as the principal mechanism among post-transcriptional and post-translational mechanisms for the dynamic regulation of gene expression.
Spirin, A.S. The second Sir Hans Krebs lecture. Informosomes. Eur. J. Biochem. 10, 20–35 (1969).
Keene, J.D. & Tenenbaum, S.A. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 9, 1161–1167 (2002).This study introduces the present conceptual model for the regulation of gene expression at the post-transcriptional level.
Candeias, M.M. et al. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat. Cell Biol. 10, 1098–1105 (2008).
Hershey, J.W., Sonenberg, N. & Mathews, M.B. Principles of translational control: an overview. Cold Spring Harb. Perspect. Biol. 4, a011528 (2012).
Shalgi, R. et al. Widespread regulation of translation by elongation pausing in heat shock. Mol. Cell 49, 439–452 (2013).
Liu, B., Han, Y. & Qian, S.B. Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol. Cell 49, 453–463 (2013).
Gerashchenko, M.V., Lobanov, A.V. & Gladyshev, V.N. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc. Natl. Acad. Sci. USA 109, 17394–17399 (2012).
Leprivier, G. et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 153, 1064–1079 (2013).
Sonenberg, N. & Hinnebusch, A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745 (2009).
Jackson, R.J., Hellen, C.U. & Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 (2010).
Baltz, A.G. et al. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell 46, 674–690 (2012).
Castello, A. et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012).
Sabatini, D.M., Erdjument-Bromage, H., Lui, M., Tempst, P. & Snyder, S.H. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 78, 35–43 (1994).
Brown, E.J. et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 369, 756–758 (1994).
Gwinn, D.M. et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 (2008).
Inoki, K., Zhu, T. & Guan, K.L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590 (2003).
Corradetti, M.N., Inoki, K., Bardeesy, N., DePinho, R.A. & Guan, K.L. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 18, 1533–1538 (2004).
Shaw, R.J. et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6, 91–99 (2004).
Hara, K. et al. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273, 14484–14494 (1998).
Wang, X., Campbell, L.E., Miller, C.M. & Proud, C.G. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem. J. 334, 261–267 (1998).
DeYoung, M.P., Horak, P., Sofer, A., Sgroi, D. & Ellisen, L.W. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14–3-3 shuttling. Genes Dev. 22, 239–251 (2008).
Brugarolas, J. et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18, 2893–2904 (2004).
Laplante, M. & Sabatini, D.M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Kim, D.H. et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 (2002).
Hara, K. et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189 (2002).
Jacinto, E. et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6, 1122–1128 (2004).
Sarbassov, D.D. et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14, 1296–1302 (2004).
Sarbassov, D.D. et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22, 159–168 (2006).
Heitman, J., Movva, N.R. & Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253, 905–909 (1991).
Gingras, A.C. et al. Regulation of 4E–BP1 phosphorylation: a novel two-step mechanism. Genes Dev. 13, 1422–1437 (1999).
Brunn, G.J. et al. Direct inhibition of the signaling functions of the mammalian target of rapamycin by the phosphoinositide 3-kinase inhibitors, wortmannin and LY294002. EMBO J. 15, 5256–5267 (1996).
Burnett, P.E., Barrow, R.K., Cohen, N.A., Snyder, S.H. & Sabatini, D.M. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E–BP1. Proc. Natl. Acad. Sci. USA 95, 1432–1437 (1998).
Hara, K. et al. Regulation of eIF-4E BP1 phosphorylation by mTOR. J. Biol. Chem. 272, 26457–26463 (1997).
Wullschleger, S., Loewith, R. & Hall, M.N. TOR signaling in growth and metabolism. Cell 124, 471–484 (2006).
Dorrello, N.V. et al. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314, 467–471 (2006).
Raught, B. et al. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 23, 1761–1769 (2004).
Pause, A. et al. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5′-cap function. Nature 371, 762–767 (1994).
Roux, P.P. & Topisirovic, I. Regulation of mRNA translation by signaling pathways. Cold Spring Harb. Perspect. Biol. 4, a012252 (2012).
Gingras, A.C. et al. Hierarchical phosphorylation of the translation inhibitor 4E–BP1. Genes Dev. 15, 2852–2864 (2001).
Zimmer, S.G., DeBenedetti, A. & Graff, J.R. Translational control of malignancy: the mRNA cap-binding protein, eIF-4E, as a central regulator of tumor formation, growth, invasion and metastasis. Anticancer Res. 20, 3A, 1343–1351 (2000).
Colina, R. et al. Translational control of the innate immune response through IRF-7. Nature 452, 323–328 (2008).This study identifies mTORC1–4E-BP–dependent translational control of IRF7, which affects the production of type 1 interferon and susceptibility to infection with vesicular stomatitis virus.
Larsson, O. et al. Apoptosis resistance downstream of eIF4E: posttranscriptional activation of an anti-apoptotic transcript carrying a consensus hairpin structure. Nucleic Acids Res. 34, 4375–4386 (2006).
Hsieh, A.C. et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012).
Thoreen, C.C. et al. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012).
Bilanges, B. et al. Tuberous sclerosis complex proteins 1 and 2 control serum-dependent translation in a TOP-dependent and -independent manner. Mol. Cell. Biol. 27, 5746–5764 (2007).
Patursky-Polischuk, I. et al. The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol. Cell. Biol. 29, 640–649 (2009).
Avni, D., Shama, S., Loreni, F. & Meyuhas, O. Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Mol. Cell. Biol. 14, 3822–3833 (1994).
Miloslavski, R. et al. Oxygen sufficiency controls TOP mRNA translation via the TSC-Rheb-mTOR pathway in a 4E-BP-independent manner. J. Mol. Cell Biol. 10.1093/jmcb/mju008 (13 March 2014).
Shama, S., Avni, D., Frederickson, R.M., Sonenberg, N. & Meyuhas, O. Overexpression of initiation factor eIF-4E does not relieve the translational repression of ribosomal protein mRNAs in quiescent cells. Gene Expr. 4, 241–252 (1995).
Damgaard, C.K. & Lykke-Andersen, J. Translational coregulation of 5′TOP mRNAs by TIA-1 and TIAR. Genes Dev. 25, 2057–2068 (2011).
Tcherkezian, J. et al. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev. 28, 357–371 (2014).
Flynn, A. & Proud, C.G. Serine 209, not serine 53, is the major site of phosphorylation in initiation factor eIF-4E in serum-treated Chinese hamster ovary cells. J. Biol. Chem. 270, 21684–21688 (1995).
Fukunaga, R. & Hunter, T. MNK1, a new MAP kinase-activated protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16, 1921–1933 (1997).
Waskiewicz, A.J., Flynn, A., Proud, C.G. & Cooper, J.A. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16, 1909–1920 (1997).
Furic, L. et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc. Natl. Acad. Sci. USA 107, 14134–14139 (2010).
Fernandez, P.C. et al. Genomic targets of the human c-Myc protein. Genes Dev. 17, 1115–1129 (2003).
Topisirovic, I. et al. Stability of eukaryotic translation initiation factor 4E mRNA is regulated by HuR, and this activity is dysregulated in cancer. Mol. Cell. Biol. 29, 1152–1162 (2009).
Cook, K.D. & Miller, J. TCR-dependent translational control of GATA-3 enhances Th2 differentiation. J. Immunol. 185, 3209–3216 (2010).
Gigoux, M. et al. Inducible costimulator facilitates T-dependent B cell activation by augmenting IL-4 translation. Mol. Immunol. 59, 46–54 (2014).
Lee, M.S., Kim, B., Oh, G.T. & Kim, Y.J. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat. Immunol. 14, 346–355 (2013).This study identifies OASL1 as a key regulator of the translation of IRF7, which affects the expression of type 1 interferon. It proposes that OASL1 acts in a negative feedback loop by suppressing the translation of IRF7 mRNA.
Bjur, E. et al. Distinct translational control in CD4+ T cell subsets. PLoS Genet. 9, e1003494 (2013).This study shows that analysis of translatomes is feasible in primary cells of the immune system that are of low abundance and that such analysis provides a perspective distinct from the analysis of their transcriptomes. It also identifies eIF4E-dependent translational control as key for the proliferation of Foxp3− and Foxp3+ CD4+ T cells.
Tangye, S.G., Ma, C.S., Brink, R. & Deenick, E.K. The good, the bad and the ugly - TFH cells in human health and disease. Nat. Rev. Immunol. 13, 412–426 (2013).
Araki, K., Ellebedy, A.H. & Ahmed, R. TOR in the immune system. Curr. Opin. Cell Biol. 23, 707–715 (2011).
Xu, H. et al. Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat. Immunol. 13, 642–650 (2012).This study shows that phosphorylation of eIF4E is key in the translational activation of IRF8 mRNA and downstream macrophage polarization.
Herdy, B. et al. Translational control of the activation of transcription factor NF-κB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nat. Immunol. 13, 543–550 (2012).
Nikolcheva, T. et al. A translational rheostat for RFLAT-1 regulates RANTES expression in T lymphocytes. J. Clin. Invest. 110, 119–126 (2002).
Krausgruber, T. et al. IRF5 promotes inflammatory macrophage polarization and TH1–TH17 responses. Nat. Immunol. 12, 231–238 (2011).
Satoh, T. et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat. Immunol. 11, 936–944 (2010).
Huang, J.T. & Schneider, R.J. Adenovirus inhibition of cellular protein synthesis involves inactivation of cap-binding protein. Cell 65, 271–280 (1991).
Kleijn, M., Vrins, C.L., Voorma, H.O. & Thomas, A.A. Phosphorylation state of the cap-binding protein eIF4E during viral infection. Virology 217, 486–494 (1996).
Connor, J.H. & Lyles, D.S. Vesicular stomatitis virus infection alters the eIF4F translation initiation complex and causes dephosphorylation of the eIF4E binding protein 4E-BP1. J. Virol. 76, 10177–10187 (2002).
Walsh, D. et al. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol. Cell. Biol. 28, 2648–2658 (2008).
Walsh, D., Perez, C., Notary, J. & Mohr, I. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J. Virol. 79, 8057–8064 (2005).
Walsh, D. & Mohr, I. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 18, 660–672 (2004).
Ben-Neriah, Y. & Karin, M. Inflammation meets cancer, with NF-κB as the matchmaker. Nat. Immunol. 12, 715–723 (2011).
Vashchenko, G. & MacGillivray, R.T. Multi-copper oxidases and human iron metabolism. Nutrients 5, 2289–2313 (2013).
Klebanoff, S.J. Bactericidal effect of Fe2+, ceruloplasmin, and phosphate. Arch. Biochem. Biophys. 295, 302–308 (1992).
Mazumder, B. & Fox, P.L. Delayed translational silencing of ceruloplasmin transcript in γ interferon-activated U937 monocytic cells: role of the 3′ untranslated region. Mol. Cell. Biol. 19, 6898–6905 (1999).
Sampath, P., Mazumder, B., Seshadri, V. & Fox, P.L. Transcript-selective translational silencing by gamma interferon is directed by a novel structural element in the ceruloplasmin mRNA 3′ untranslated region. Mol. Cell. Biol. 23, 1509–1519 (2003).
Vyas, K. et al. Genome-wide polysome profiling reveals an inflammation-responsive posttranscriptional operon in gamma interferon-activated monocytes. Mol. Cell. Biol. 29, 458–470 (2009).
Mukhopadhyay, R. et al. DAPK-ZIPK-L13a axis constitutes a negative-feedback module regulating inflammatory gene expression. Mol. Cell 32, 371–382 (2008).These authors identify a negative feedback loop for GAIT complex activity. DAPK and ZIPK, which are activators of the GAIT complex, are themselves targets for suppressed translation via a GAIT-dependent mechanism.
Scheu, S. et al. Activation of the integrated stress response during T helper cell differentiation. Nat. Immunol. 7, 644–651 (2006).
Wek, R.C., Jiang, H.Y. & Anthony, T.G. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34, 7–11 (2006).
Villarino, A.V. et al. Posttranscriptional silencing of effector cytokine mRNA underlies the anergic phenotype of self-reactive T cells. Immunity 34, 50–60 (2011).This study shows that translational control of cytokines is important for self reactive T-cell anergy.
Piecyk, M. et al. TIA-1 is a translational silencer that selectively regulates the expression of TNF-α. EMBO J. 19, 4154–4163 (2000).
Katsanou, V. et al. HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell 19, 777–789 (2005).
Yu, C. et al. An essential function of the SRC-3 coactivator in suppression of cytokine mRNA translation and inflammatory response. Mol. Cell 25, 765–778 (2007).
Dixon, D.A. et al. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J. Exp. Med. 198, 475–481 (2003).
Dhamija, S. et al. Interleukin-17 (IL-17) and IL-1 activate translation of overlapping sets of mRNAs, including that of the negative regulator of inflammation, MCPIP1. J. Biol. Chem. 288, 19250–19259 (2013).
Yamamoto, M. et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IκBζ. Nature 430, 218–222 (2004).
Chang, C.H. et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013).
Warner, J.R., Knopf, P.M. & Rich, A. A multiple ribosomal structure in protein synthesis. Proc. Natl. Acad. Sci. USA 49, 122–129 (1963).
Johannes, G., Carter, M.S., Eisen, M.B., Brown, P.O. & Sarnow, P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. USA 96, 13118–13123 (1999).
Karginov, F.V. & Hannon, G.J. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 27, 1624–1632 (2013).
Ingolia, N.T., Ghaemmaghami, S., Newman, J.R. & Weissman, J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).This study introduces the ribosome-profiling technique as a genome-wide tool with which to map the positions of ribosomes on mRNA.
Ingolia, N.T., Brar, G.A., Rouskin, S., McGeachy, A.M. & Weissman, J.S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534–1550 (2012).
Zoschke, R., Watkins, K.P. & Barkan, A. A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell 25, 2265–2275 (2013).
Larsson, O., Sonenberg, N. & Nadon, R. Identification of differential translation in genome wide studies. Proc. Natl. Acad. Sci. USA 107, 21487–21492 (2010).
Olshen, A.B. et al. Assessing gene-level translational control from ribosome profiling. Bioinformatics 29, 2995–3002 (2013).
Larsson, O & Nadon, R. Re-analysis of genome wide data on mammalian microRNA-mediated suppression of gene expression. Translation 1, 1–9 (2013).
Eliseeva, I.A., Vorontsov, I.E., Babeyev, K.E., Buyanova, S.M., Sysoeva, M.A. & Kondrashov, F.A. et al. In silico motif analysis suggests an interplay of transcriptional and translational control in mTOR response. Translation 1, 1–7 (2013).
Larsson, O., Sonenberg, N. & Nadon, R. Anota: Analysis of differential translation in genome-wide studies. Bioinformatics 27, 1440–1441 (2011).
Colman, H. et al. Genome-wide analysis of host mRNA translation during hepatitis C virus infection. J. Virol. 87, 6668–6677 (2013).This article shows that the analysis approach (Anota or translational-efficiency score) is critical for predicting whether differences in translation identified will correlate with changes in proteomes. Only Anota analysis corresponds to changes in protein amounts.
Larsson, O. et al. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proc. Natl. Acad. Sci. USA 109, 8977–8982 (2012).
Parker, M.W. et al. Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J. Clin. Invest. 124, 1622–1635 (2014).
Stumpf, C.R., Moreno, M.V., Olshen, A.B., Taylor, B.S. & Ruggero, D. The translational landscape of the mammalian cell cycle. Mol. Cell 52, 574–582 (2013).
Tebaldi, T., Dassi, E., Kostoska, G., Viero, G. & Quattrone, A. tRanslatome: an R/Bioconductor package to portray translational control. Bioinformatics 30, 289–291 (2014).
Michel, A.M. et al. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res. 42, D859–D864 (2014).
Schneider-Poetsch, T. et al. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6, 209–217 (2010).
Lee, S. et al. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc. Natl. Acad. Sci. USA 109, E2424–E2432 (2012).
Ingolia, N.T., Lareau, L.F. & Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of Mammalian proteomes. Cell 147, 789–802 (2011).
Guttman, M., Russell, P., Ingolia, N.T., Weissman, J.S. & Lander, E.S. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154, 240–251 (2013).
Fresno, M., Jimenez, A. & Vazquez, D. Inhibition of translation in eukaryotic systems by harringtonine. Eur. J. Biochem. 72, 323–330 (1977).
Larsson, O. et al. Eukaryotic translation initiation factor 4E induced progression of primary human mammary epithelial cells along the cancer pathway is associated with targeted translational deregulation of oncogenic drivers and inhibitors. Cancer Res. 67, 6814–6824 (2007).
Mamane, Y. et al. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PLoS ONE 2, e242 (2007).
Rajasekhar, V.K. et al. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol. Cell 12, 889–901 (2003).
Tominaga, Y., Tamguney, T., Kolesnichenko, M., Bilanges, B. & Stokoe, D. Translational deregulation in PDK-1−/− embryonic stem cells. Mol. Cell. Biol. 25, 8465–8475 (2005).
Mikulits, W. et al. Isolation of translationally controlled mRNAs by differential screening. FASEB J. 14, 1641–1652 (2000).
Grolleau, A. et al. Global and specific translational control by rapamycin in T cells uncovered by microarrays and proteomics. J. Biol. Chem. 277, 22175–22184 (2002).
Ceppi, M. et al. Ribosomal protein mRNAs are translationally-regulated during human dendritic cells activation by LPS. Immunome Res. 5, 5 (2009).
Kitamura, H. et al. Genome-wide identification and characterization of transcripts translationally regulated by bacterial lipopolysaccharide in macrophage-like J774.1 cells. Physiol. Genomics 33, 121–132 (2008).
Ring, A.M. et al. Mechanistic and structural insight into the functional dichotomy between IL-2 and IL-15. Nat. Immunol. 13, 1187–1195 (2012).
Acknowledgements
Supported by the Canadian Institutes for Health Research (I.T., and MOP 67211 to C.A.P.), the Canada Research Chair program (C.A.P.), the Swedish Research Council (O.L.), the Swedish Cancer Society (O.L.) and the Wallenberg Academy Fellows program (O.L.).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Piccirillo, C., Bjur, E., Topisirovic, I. et al. Translational control of immune responses: from transcripts to translatomes. Nat Immunol 15, 503–511 (2014). https://doi.org/10.1038/ni.2891
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2891
This article is cited by
-
Translational profiling identifies sex-specific metabolic and epigenetic reprogramming of cortical microglia/macrophages in APPPS1-21 mice with an antibiotic-perturbed-microbiome
Molecular Neurodegeneration (2023)
-
25KDa branched polyethylenimine increases interferon-γ production in natural killer cells via improving translation efficiency
Cell Communication and Signaling (2023)
-
The Akt/mTOR and MNK/eIF4E pathways rewire the prostate cancer translatome to secrete HGF, SPP1 and BGN and recruit suppressive myeloid cells
Nature Cancer (2023)
-
Protein synthesis, degradation, and energy metabolism in T cell immunity
Cellular & Molecular Immunology (2022)
-
High expression of eIF4E is associated with tumor macrophage infiltration and leads to poor prognosis in breast cancer
BMC Cancer (2021)