Abstract
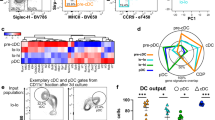
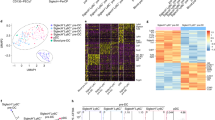
Monocytes, macrophages and dendritic cells (DCs) are developmentally related regulators of the immune system that share the monocyte-macrophage DC progenitor (MDP) as a common precursor. Unlike differentiation into DCs, the distal pathways for differentiation into monocytes and monocyte-derived macrophages are not fully elucidated. We have now demonstrated the existence of a clonogenic, monocyte- and macrophage-restricted progenitor cell derived from the MDP. This progenitor was a Ly6C+ proliferating cell present in the bone marrow and spleen that generated the major monocyte subsets and macrophages, but not DCs or neutrophils. By in-depth quantitative proteomics, we characterized changes in the proteome during monocyte differentiation, which provided insight into the molecular principles of developing monocytes, such as their functional maturation. Thus, we found that monocytes and macrophages were renewed independently of DCs from a committed progenitor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
van Furth, R. & Cohn, Z.A. The origin and kinetics of mononuclear phagocytes. J. Exp. Med. 128, 415–435 (1968).
Auffray, C., Sieweke, M.H. & Geissmann, F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu. Rev. Immunol. 27, 669–692 (2009).
Serbina, N.V., Jia, T., Hohl, T.M. & Pamer, E.G. Monocyte-mediated defense against microbial pathogens. Annu. Rev. Immunol. 26, 421–452 (2008).
Tacke, F. et al. Monocyte subsets differentially employ CCR2, CCR5, and CX3CR1 to accumulate within atherosclerotic plaques. J. Clin. Invest. 117, 185–194 (2007).
Swirski, F.K. et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science 325, 612–616 (2009).
Martin, A.P. et al. Increased expression of CCL2 in insulin-producing cells of transgenic mice promotes mobilization of myeloid cells from the bone marrow, marked insulitis, and diabetes. Diabetes 57, 3025–3033 (2008).
Qian, B.Z. et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 475, 222–225 (2011).
Geissmann, F., Jung, S. & Littman, D.R. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity 19, 71–82 (2003).
Passlick, B., Flieger, D. & Ziegler-Heitbrock, H.W. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood 74, 2527–2534 (1989).
Ingersoll, M.A. et al. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood 115, e10–e19 (2010).
Ziegler-Heitbrock, L. et al. Nomenclature of monocytes and dendritic cells in blood. Blood 116, e74–e80 (2010).
Geissmann, F. et al. Development of monocytes, macrophages, and dendritic cells. Science 327, 656–661 (2010).
Auffray, C. et al. Monitoring of blood vessels and tissues by a population of monocytes with patrolling behavior. Science 317, 666–670 (2007).
Schulz, C. et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86–90 (2012).
Yona, S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2012).
Ginhoux, F. et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010).
Hoeffel, G. et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J. Exp. Med. 209, 1167–1181 (2012).
Cheong, C. et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209+ dendritic cells for immune T cell areas. Cell 143, 416–429 (2010).
van Furth, R., Hirsch, J.G. & Fedorko, M.E. Morphology and peroxidase cytochemistry of mouse promonocytes, monocytes, and macrophages. J. Exp. Med. 132, 794–812 (1970).
Goud, T.J., Schotte, C. & van Furth, R. Identification and characterization of the monoblast in mononuclear phagocyte colonies grown in vitro. J. Exp. Med. 142, 1180–1199 (1975).
Geissmann, F., Gordon, S., Hume, D.A., Mowat, A.M. & Randolph, G.J. Unravelling mononuclear phagocyte heterogeneity. Nat. Rev. Immunol. 10, 453–460 (2010).
Fogg, D.K. et al. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science 311, 83–87 (2006).
Auffray, C. et al. CX3CR1+CD115+CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J. Exp. Med. 206, 595–606 (2009).
Onai, N. et al. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8, 1207–1216 (2007).
Naik, S.H. et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8, 1217–1226 (2007).
Liu, K. et al. In vivo analysis of dendritic cell development and homeostasis. Science 324, 392–397 (2009).
Saha, P. & Geissmann, F. Toward a functional characterization of blood monocytes. Immunol. Cell Biol. 89, 2–4 (2011).
Merad, M. & Manz, M.G. Dendritic cell homeostasis. Blood 113, 3418–3427 (2009).
Waskow, C. et al. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat. Immunol. 9, 676–683 (2008).
Kiel, M.J. & Morrison, S.J. Uncertainty in the niches that maintain haematopoietic stem cells. Nat. Rev. Immunol. 8, 290–301 (2008).
Leuschner, F. et al. Rapid monocyte kinetics in acute myocardial infarction are sustained by extramedullary monocytopoiesis. J. Exp. Med. 209, 123–137 (2012).
Sallusto, F. & Lanzavecchia, A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor α. J. Exp. Med. 179, 1109–1118 (1994).
Akashi, K., Traver, D., Miyamoto, T. & Weissman, I.L. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404, 193–197 (2000).
Varol, C. et al. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J. Exp. Med. 204, 171–180 (2007).
Heng, T.S. & Painter, M.W. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008).
Sardiello, M. et al. A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009).
Krausgruber, T. et al. IRF5 promotes inflammatory macrophage polarization and TH1–TH17 responses. Nat. Immunol. 12, 231–238 (2011).
Moran, A.E. et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208, 1279–1289 (2011).
Hanna, R.N. et al. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C− monocytes. Nat. Immunol. 12, 778–785 (2011).
Sunderkötter, C. et al. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J. Immunol. 172, 4410–4417 (2004).
Ancuta, P. et al. Transcriptional profiling reveals developmental relationship and distinct biological functions of CD16+ and CD16− monocyte subsets. BMC Genomics 10, 403 (2009).
Leuschner, F. et al. Therapeutic siRNA silencing in inflammatory monocytes in mice. Nat. Biotechnol. 29, 1005–1010 (2011).
Garcia, M.R. et al. Monocytic suppressive cells mediate cardiovascular transplantation tolerance in mice. J. Clin. Invest. 120, 2486–2496 (2010).
Miller, J.C. et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13, 888–899 (2012).
Shen, F.W. et al. Cloning of Ly-5 cDNA. Proc. Natl. Acad. Sci. USA 82, 7360–7363 (1985).
Kim, J.M., Rasmussen, J.P. & Rudensky, A.Y. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat. Immunol. 8, 191–197 (2007).
Jung, S. et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell Biol. 20, 4106–4114 (2000).
Boersema, P.J., Raijmakers, R., Lemeer, S., Mohammed, S. & Heck, A.J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 (2009).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Nikolsky, Y., Ekins, S., Nikolskaya, T. & Bugrim, A. A novel method for generation of signature networks as biomarkers from complex high throughput data. Toxicol. Lett. 158, 20–29 (2005).
Acknowledgements
We thank A. Rudensky (Memorial Sloan-Kettering Cancer Center, New York) for B6N.129(Cg)-Foxp3tm3Ayr mice; M. Platten (German Cancer Research Center) for Cx3cr1-eGFP reporter mice; F. Rosenbauer and S. Ghani for help with the in vitro differentiation assay; the Central Animal Laboratory, Flow Cytometry and Light Microscopy core facilities of the German Cancer Research Center, as well as the European Molecular Biology Laboratory Proteomics Core Facility, for training and support; H.R. Rodewald, T. Feyerabend and B. Kyewski for support and discussions; P. Pyl for statistical advice; K. Lobbes, A. Schmälzle and C. Maul for technical support; and the Immunological Genome Project Consortium for assembled data35,44. Supported by the Helmholtz Association of German Research Centers (J.He. and M.M.B.; and HZ-NG-505 to M.F.) and the Netherlands Organization for Scientific Research (to J.K.).
Author information
Authors and Affiliations
Contributions
J.He., D.M.R., J.Ha., J.K. and M.F. designed experiments; J.He., D.M.R., J.Ha., M.M.B. and A.-C.J. did experiments; J.Ha. and J.K. did mass spectrometry; and J.He., D.M.R., J.Ha., M.M.B., J.K. and M.F. analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures and Tables
Supplementary Figures 1–8 and Supplementary Tables 1, 2, 5, 6 and 7 (PDF 2043 kb)
Supplementary Table 3
Full proteomics table for a total of 5967 proteins identified in the cMoP, MDP and Ly6Chigh monocyte proteome (XLSX 5235 kb)
Supplementary Table 4
GO process enrichment for comparison of the cMoP (upper part) and Ly6Chigh monocytes (lower part) (XLSX 39 kb)
Rights and permissions
About this article
Cite this article
Hettinger, J., Richards, D., Hansson, J. et al. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol 14, 821–830 (2013). https://doi.org/10.1038/ni.2638
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2638
This article is cited by
-
Advances in lung ischemia/reperfusion injury: unraveling the role of innate immunity
Inflammation Research (2024)
-
Spleen regeneration after subcutaneous heterotopic autotransplantation in a mouse model
Biological Research (2023)
-
Specific inflammatory osteoclast precursors induced during chronic inflammation give rise to highly active osteoclasts associated with inflammatory bone loss
Bone Research (2022)
-
Ablation of cDC2 development by triple mutations within the Zeb2 enhancer
Nature (2022)
-
Neutrophils in cancer: heterogeneous and multifaceted
Nature Reviews Immunology (2022)