Abstract
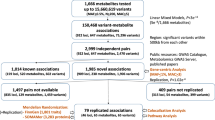
Genetic factors modifying the blood metabolome have been investigated through genome-wide association studies (GWAS) of common genetic variants and through exome sequencing. We conducted a whole-genome sequencing study of common, low-frequency and rare variants to associate genetic variations with blood metabolite levels using comprehensive metabolite profiling in 1,960 adults. We focused the analysis on 644 metabolites with consistent levels across three longitudinal data collections. Genetic sequence variations at 101 loci were associated with the levels of 246 (38%) metabolites (P ≤ 1.9 × 10−11). We identified 113 (10.7%) among 1,054 unrelated individuals in the cohort who carried heterozygous rare variants likely influencing the function of 17 genes. Thirteen of the 17 genes are associated with inborn errors of metabolism or other pediatric genetic conditions. This study extends the map of loci influencing the metabolome and highlights the importance of heterozygous rare variants in determining abnormal blood metabolic phenotypes in adults.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Yousri, N.A. et al. Long term conservation of human metabolic phenotypes and link to heritability. Metabolomics 10, 1005–1017 (2014).
Suhre, K. & Gieger, C. Genetic variation in metabolic phenotypes: study designs and applications. Nat. Rev. Genet. 13, 759–769 (2012).
Kastenmüller, G., Raffler, J., Gieger, C. & Suhre, K. Genetics of human metabolism: an update. Hum. Mol. Genet. 24, R93–R101 (2015).
Suhre, K. et al. A genome-wide association study of metabolic traits in human urine. Nat. Genet. 43, 565–569 (2011).
Shin, S.Y. et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 46, 543–550 (2014).
Draisma, H.H. et al. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat. Commun. 6, 7208 (2015).
Kettunen, J. et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat. Commun. 7, 11122 (2016).
Guo, L. et al. Plasma metabolomic profiles enhance precision medicine for volunteers of normal health. Proc. Natl. Acad. Sci. USA 112, E4901–E4910 (2015).
Yu, B. et al. Association of rare loss-of-function alleles in HAL, serum histidine: levels and incident coronary heart disease. Circ Cardiovasc Genet 8, 351–355 (2015).
Yazdani, A., Yazdani, A., Liu, X. & Boerwinkle, E. Identification of rare variants in metabolites of the carnitine pathway by whole genome sequencing analysis. Genet. Epidemiol. 40, 486–491 (2016).
Rhee, E.P. et al. An exome array study of the plasma metabolome. Nat. Commun. 7, 12360 (2016).
Moayyeri, A., Hammond, C.J., Hart, D.J. & Spector, T.D. The UK Adult Twin Registry (TwinsUK Resource). Twin Res. Hum. Genet. 16, 144–149 (2013).
Moayyeri, A., Hammond, C.J., Valdes, A.M. & Spector, T.D. Cohort profile: TwinsUK and healthy ageing twin study. Int. J. Epidemiol. 42, 76–85 (2013).
Telenti, A. et al. Deep sequencing of 10,000 human genomes. Proc. Natl. Acad. Sci. USA 113, 11901–11906 (2016).
Xu, C. et al. Estimating genome-wide significance for whole-genome sequencing studies. Genet. Epidemiol. 38, 281–290 (2014).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015).
Demirkan, A. et al. Insight in genome-wide association of metabolite quantitative traits by exome sequence analyses. PLoS Genet. 11, e1004835 (2015).
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP–trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
Krumsiek, J. et al. Mining the unknown: a systems approach to metabolite identification combining genetic and metabolic information. PLoS Genet. 8, e1003005 (2012).
Wu, M.C. et al. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 89, 82–93 (2011).
Pedersen, C.B. et al. The ACADS gene variation spectrum in 114 patients with short-chain acyl-CoA dehydrogenase (SCAD) deficiency is dominated by missense variations leading to protein misfolding at the cellular level. Hum. Genet. 124, 43–56 (2008).
Goodman, S.I., Binard, R.J., Woontner, M.R. & Frerman, F.E. Glutaric acidemia type II: gene structure and mutations of the electron transfer flavoprotein:ubiquinone oxidoreductase (ETF:QO) gene. Mol. Genet. Metab. 77, 86–90 (2002).
Ionita-Laza, I., McCallum, K., Xu, B. & Buxbaum, J.D. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 48, 214–220 (2016).
Kuokkanen, M. et al. Mutations in the translated region of the lactase gene (LCT) underlie congenital lactase deficiency. Am. J. Hum. Genet. 78, 339–344 (2006).
Enattah, N.S. et al. Identification of a variant associated with adult-type hypolactasia. Nat. Genet. 30, 233–237 (2002).
McGill, J.B. et al. Circulating 1,5-anhydroglucitol levels in adult patients with diabetes reflect longitudinal changes of glycemia: a U.S. trial of the GlycoMark assay. Diabetes Care 27, 1859–1865 (2004).
Koga, M., Murai, J., Saito, H., Mukai, M. & Kasayama, S. Habitual intake of dairy products influences serum 1,5-anhydroglucitol levels independently of plasma glucose. Diabetes Res. Clin. Pract. 90, 122–125 (2010).
Yamanouchi, T. et al. Common reabsorption system of 1,5-anhydro-D-glucitol, fructose, and mannose in rat renal tubule. Biochim. Biophys. Acta 1291, 89–95 (1996).
Grempler, R. et al. Functional characterisation of human SGLT-5 as a novel kidney-specific sodium-dependent sugar transporter. FEBS Lett. 586, 248–253 (2012).
Dworacka, M. et al. 1,5-anhydro-D-glucitol: a novel marker of glucose excursions. Int. J. Clin. Pract. Suppl. 129, 40–44 (2002).
Her, C. et al. Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics 53, 284–295 (1998).
Gregersen, N. et al. Identification of four new mutations in the short-chain acyl-CoA dehydrogenase (SCAD) gene in two patients: one of the variant alleles, 511C→T, is present at an unexpectedly high frequency in the general population, as was the case for 625G→A, together conferring susceptibility to ethylmalonic aciduria. Hum. Mol. Genet. 7, 619–627 (1998).
Goldstein, D.S. et al. Sources and physiological significance of plasma dopamine sulfate. J. Clin. Endocrinol. Metab. 84, 2523–2531 (1999).
Suhre, K. et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 477, 54–60 (2011).
Gieger, C. et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 4, e1000282 (2008).
Cirulli, E.T. & Goldstein, D.B. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat. Rev. Genet. 11, 415–425 (2010).
Menni, C. et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol. 42, 1111–1119 (2013).
Schwenger, B., Schober, S. & Simon, D. DUMPS cattle carry a point mutation in the uridine monophosphate synthase gene. Genomics 16, 241–244 (1993).
Imaeda, M. et al. Hereditary orotic aciduria heterozygotes accompanied with neurological symptoms. Tohoku J. Exp. Med. 185, 67–70 (1998).
Corydon, M.J. et al. Role of common gene variations in the molecular pathogenesis of short-chain acyl-CoA dehydrogenase deficiency. Pediatr. Res. 49, 18–23 (2001).
Béhin, A. et al. Multiple acyl-CoA dehydrogenase deficiency (MADD) as a cause of late-onset treatable metabolic disease. Rev. Neurol. (Paris) 172, 231–241 (2016).
Visscher, P.M., Benyamin, B. & White, I. The use of linear mixed models to estimate variance components from data on twin pairs by maximum likelihood. Twin Res. 7, 670–674 (2004).
Scheike, T.H., Holst, K.K. & Hjelmborg, J.B. Estimating heritability for cause specific mortality based on twin studies. Lifetime Data Anal. 20, 210–233 (2014).
Raczy, C. et al. Isaac: ultra-fast whole-genome secondary analysis on Illumina sequencing platforms. Bioinformatics 29, 2041–2043 (2013).
Alexander, D.H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Moltke, I. & Albrechtsen, A. RelateAdmix: a software tool for estimating relatedness between admixed individuals. Bioinformatics 30, 1027–1028 (2014).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Yu, J. et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 38, 203–208 (2006).
Widmer, C. et al. Further improvements to linear mixed models for genome-wide association studies. Sci. Rep. 4, 6874 (2014).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92 (2012).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Yates, A. et al. Ensembl 2016. Nucleic Acids Res. 44, D710–D716 (2016).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Govindasamy, L. et al. Structural and mutational characterization of L-carnitine binding to human carnitine acetyltransferase. J. Struct. Biol. 146, 416–424 (2004).
Heinrich, D., Diederichsen, U. & Rudolph, M.G. Lys314 is a nucleophile in non-classical reactions of orotidine-5′-monophosphate decarboxylase. Chemistry 15, 6619–6625 (2009).
Lee, K.A. et al. Crystal structure of human cholesterol sulfotransferase (SULT2B1b) in the presence of pregnenolone and 3′-phosphoadenosine 5′-phosphate. Rationale for specificity differences between prototypical SULT2A1 and the SULT2BG1 isoforms. J. Biol. Chem. 278, 44593–44599 (2003).
Kiefer, F., Arnold, K., Kunzli, M., Bordoli, L. & Schwede, T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 37, D387–D392 (2009).
Kopp, J. & Schwede, T. The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 32, D230–D234 (2004).
Pieper, U. et al. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 32, D217–D222 (2004).
Acknowledgements
The sequencing and metabolome study was funded by Human Longevity, Inc. TwinsUK was funded by the Wellcome Trust, European Community's Seventh Framework Programme (FP7/2007-2013 277849, 201413 and 259749). The study also receives support from the National Institute for Health Research (NIHR) Clinical Research Facility at Guy's and St Thomas' NHS Foundation Trust and the NIHR Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London. T.D.S. is an NIHR senior Investigator.
Author information
Authors and Affiliations
Contributions
J.C.V. conceived the study. A.T. led the analyses. T.L. and A.T. designed the study. T.L. performed genome analyses. M.H. performed structural analysis. H.-C.Y. performed Mendelian and pathway analyses. W.H.B. led the sequencing process. C.M., J.Z., K.S., M.M. and T.D.S. are responsible for the Twin Cohort study. A.M.E., L.A.D.M. and L.G. contributed metabolome expertise and data. H.M., B.A.P. and C.T.C. contributed clinical support. E.F.K., S.B., Y.T., N.J.S. and C.G. supervised research.
Corresponding authors
Ethics declarations
Competing interests
The following authors are current employees or stockholders of Human Longevity, Inc.: J.C.V., A.T., T.L., M.H., H.-C.Y., W.H.B., E.F.K., S.B., Y.T., B.A.P. and N.J.S. The following authors are current employees or stockholders of Metabolome, Inc.: A.M.E., L.A.D.M. and L.G.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–16 (PDF 7521 kb)
Supplementary Table 1
Metabolite h2 and outliers. (XLSX 166 kb)
Supplementary Table 2
GWAS significant independent variants. (XLSX 100 kb)
Supplementary Table 3
GWAS summary statistics. (XLSX 19696 kb)
Supplementary Table 4
GWAS comparison. (XLSX 104 kb)
Supplementary Table 5
SKAT results. (XLSX 155 kb)
Supplementary Table 6
Additional coding rare variants. (XLSX 32 kb)
Supplementary Table 7
Promoter rare variants in outliers. (XLSX 28 kb)
Supplementary Table 8
Rare variants from publications. (XLSX 30 kb)
Supplementary Table 9
Rare variants in genes from publications. (XLSX 30 kb)
Rights and permissions
About this article
Cite this article
Long, T., Hicks, M., Yu, HC. et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat Genet 49, 568–578 (2017). https://doi.org/10.1038/ng.3809
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3809
This article is cited by
-
Identification of blood metabolites associated with risk of Alzheimer’s disease by integrating genomics and metabolomics data
Molecular Psychiatry (2024)
-
Genetic influences on circulating retinol and its relationship to human health
Nature Communications (2024)
-
Genome-wide characterization of circulating metabolic biomarkers
Nature (2024)
-
Plasma metabolomics profiles in Black and White participants of the Adventist Health Study-2 cohort
BMC Medicine (2023)
-
Ancestry-driven metabolite variation provides insights into disease states in admixed populations
Genome Medicine (2023)