Abstract
Wilms tumor is the most common childhood renal cancer1. To identify mutations that predispose to Wilms tumor, we are conducting exome sequencing studies. Here we describe 11 different inactivating mutations in the REST gene (encoding RE1-silencing transcription factor) in four familial Wilms tumor pedigrees and nine non-familial cases. Notably, no similar mutations were identified in the ICR1000 control series2 (13/558 versus 0/993; P < 0.0001) or in the ExAC series (13/558 versus 0/61,312; P < 0.0001). We identified a second mutational event in two tumors, suggesting that REST may act as a tumor-suppressor gene in Wilms tumor pathogenesis. REST is a zinc-finger transcription factor that functions in cellular differentiation and embryonic development3,4. Notably, ten of 11 mutations clustered within the portion of REST encoding the DNA-binding domain, and functional analyses showed that these mutations compromise REST transcriptional repression. These data establish REST as a Wilms tumor predisposition gene accounting for ∼2% of Wilms tumor.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
08 February 2016
In the version of this article initially published, the authors failed to acknowledge funding from the NIHR Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and University College London to Neil Sebire. The error has been corrected in the HTML and PDF versions of the article.
References
Hohenstein, P., Pritchard-Jones, K. & Charlton, J. The yin and yang of kidney development and Wilms' tumors. Genes Dev. 29, 467–482 (2015).
Ruark, E. et al. The ICR1000 UK exome series: a resource of gene variation in an outbred population. F1000 Res. 4, 883 (2015).
Bithell, A. REST: transcriptional and epigenetic regulator. Epigenomics 3, 47–58 (2011).
Ooi, L. & Wood, I.C. Chromatin crosstalk in development and disease: lessons from REST. Nat. Rev. Genet. 8, 544–554 (2007).
Scott, R.H., Stiller, C.A., Walker, L. & Rahman, N. Syndromes and constitutional chromosomal abnormalities associated with Wilms tumour. J. Med. Genet. 43, 705–715 (2006).
Hanks, S. et al. Germline mutations in the PAF1 complex gene CTR9 predispose to Wilms tumour. Nat. Commun. 5, 4398 (2014).
Rahman, N. et al. Evidence for a familial Wilms' tumour gene (FWT1) on chromosome 17q12-q21. Nat. Genet. 13, 461–463 (1996).
McDonald, J.M. et al. Linkage of familial Wilms' tumor predisposition to chromosome 19 and a two-locus model for the etiology of familial tumors. Cancer Res. 58, 1387–1390 (1998).
Rapley, E.A. et al. Evidence for susceptibility genes to familial Wilms tumour in addition to WT1, FWT1 and FWT2. Br. J. Cancer 83, 177–183 (2000).
Ruark, E. et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature 493, 406–410 (2013).
Forbes, S.A. et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 39, D945–D950 (2011).
Lu, T. et al. REST and stress resistance in ageing and Alzheimer's disease. Nature 507, 448–454 (2014).
Buckley, N.J., Johnson, R., Zuccato, C., Bithell, A. & Cattaneo, E. The role of REST in transcriptional and epigenetic dysregulation in Huntington's disease. Neurobiol. Dis. 39, 28–39 (2010).
Canzonetta, C. et al. DYRK1A-dosage imbalance perturbs NRSF/REST levels, deregulating pluripotency and embryonic stem cell fate in Down syndrome. Am. J. Hum. Genet. 83, 388–400 (2008).
Karlin, K.L. et al. The oncogenic STP axis promotes triple-negative breast cancer via degradation of the REST tumor suppressor. Cell Rep. 9, 1318–1332 (2014).
Gurrola-Diaz, C., Lacroix, J., Dihlmann, S., Becker, C.M. & von Knebel Doeberitz, M. Reduced expression of the neuron restrictive silencer factor permits transcription of glycine receptor α1 subunit in small-cell lung cancer cells. Oncogene 22, 5636–5645 (2003).
Kreisler, A. et al. Regulation of the NRSF/REST gene by methylation and CREB affects the cellular phenotype of small-cell lung cancer. Oncogene 29, 5828–5838 (2010).
Tawadros, T. et al. IB1/JIP-1 controls JNK activation and increased during prostatic LNCaP cells neuroendocrine differentiation. Cell. Signal. 17, 929–939 (2005).
Ballas, N., Grunseich, C., Lu, D.D., Speh, J.C. & Mandel, G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121, 645–657 (2005).
Little, S.E. et al. Frequency and heritability of WT1 mutations in nonsyndromic Wilms' tumor patients: a UK Children's Cancer Study Group Study. J. Clin. Oncol. 22, 4140–4146 (2004).
Scott, R.H. et al. Surveillance for Wilms tumour in at-risk children: pragmatic recommendations for best practice. Arch. Dis. Child. 91, 995–999 (2006).
Lunter, G. & Goodson, M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21, 936–939 (2011).
Rimmer, A. et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 46, 912–918 (2014).
Acknowledgements
We thank the families for their participation in our research and the physicians and nurses who recruited them. Samples were collected through the Factors Associated with Childhood Tumours (FACT) study, which is a Children's Cancer and Leukaemia (CCLG) Study (UK National Research Ethics Service reference 05/MRE02/17). The individuals who recruited patients with Wilms tumor for this study are listed in the Supplementary Note. We thank M. Warren-Perry and J. Bull for assistance in recruitment and A. Strydom for assistance in preparing the manuscript. We acknowledge NHS funding to the Royal Marsden/Institute of Cancer Research National Institute for Health Research (NIHR) Biomedical Research Centre. The authors acknowledge joint participation by the Adrienne Helis Melvin Medical Research Foundation through its direct engagement in the continuous active conduct of medical research in conjunction with Baylor College of Medicine for cancer research. T.F.W. and K.L.K. were supported by the Cancer Prevention Research Institute of Texas (CPRIT; RP120583), the US National Institutes of Health (1R01CA178039-01) and the US Department of Defense Breast Cancer Research Program (BC120604). This research was supported by the Wellcome Trust (088804/Z/09/Z) and the Rosetrees Trust. This research was supported by the NIHR Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and University College London (to N.S.).
Author information
Authors and Affiliations
Contributions
N.R. designed and oversaw the study. S.S.M., S.H., E.R.P., Anthony Renwick, E. Ramsay and S.S. performed the molecular analyses. E. Ruark, A.E., S.Y., C.A.S. and Alexander Renwick performed bioinformatics analyses. J.B., M.C., J.G., J.H., J.K., G.L., T.M., E.S. and C.S. provided samples and data, coordinated by A.Z. K.L.K. and T.F.W. undertook functional analyses. N.S. undertook pathology review. S.S.M., S.H., K.L.K., T.F.W. and N.R. wrote the manuscript with input from the other authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
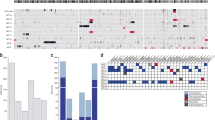
Supplementary Figure 2 Pedigrees of Wilms tumor cases with germline REST mutations.
Individuals with Wilms tumor (WT) are represented by shaded symbols, with age at diagnosis in years given below. Circles depict females, and squares depict males.
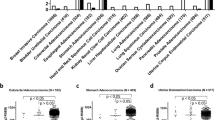
Supplementary Figure 3 REST mutants identified in Wilms tumor cases are deficient in transcriptional repression relative to the normalizing control GAPDH.
(a) Expression of REST target genes was assessed in REST-deficient breast cancer cells (SUM149) expressing the indicated cDNAs by qRT-PCR. Relative expression of the REST target complexin 1 is not shown, as it is not expressed in the SUM149 cell line. (b) Expression of REST target genes was assessed in REST-deficient colon cancer cells (SW1417) expressing the indicated cDNAs by qRT-PCR. GFP, green fluorescent protein control; WT, wild-type REST. REST mutations within the DNA-binding domain and/or truncating mutations are shown in bold.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Tables 1–3 and Supplementary Note. (PDF 2507 kb)
Rights and permissions
About this article
Cite this article
Mahamdallie, S., Hanks, S., Karlin, K. et al. Mutations in the transcriptional repressor REST predispose to Wilms tumor. Nat Genet 47, 1471–1474 (2015). https://doi.org/10.1038/ng.3440
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3440
This article is cited by
-
Hallmark discoveries in the biology of Wilms tumour
Nature Reviews Urology (2024)
-
Pathogenic REST variant causing Jones syndrome and a review of the literature
European Journal of Human Genetics (2023)
-
The cause of Jones syndrome put to REST: a mutation in the REST gene causes gingival fibromatosis and hearing loss
European Journal of Human Genetics (2023)
-
Differential expression profiling of onco and tumor-suppressor genes from major-signaling pathways in Wilms’ tumor
Pediatric Surgery International (2022)
-
The genetic changes of Wilms tumour
Nature Reviews Nephrology (2019)