Abstract
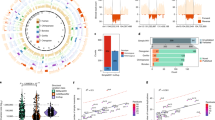
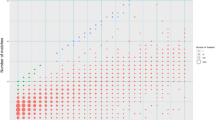
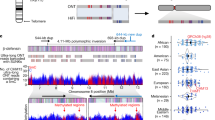
Recurrent deletions of chromosome 15q13.3 associate with intellectual disability, schizophrenia, autism and epilepsy. To gain insight into the instability of this region, we sequenced it in affected individuals, normal individuals and nonhuman primates. We discovered five structural configurations of the human chromosome 15q13.3 region ranging in size from 2 to 3 Mb. These configurations arose recently (∼0.5–0.9 million years ago) as a result of human-specific expansions of segmental duplications and two independent inversion events. All inversion breakpoints map near GOLGA8 core duplicons—a ∼14-kb primate-specific chromosome 15 repeat that became organized into larger palindromic structures. GOLGA8-flanked palindromes also demarcate the breakpoints of recurrent 15q13.3 microdeletions, the expansion of chromosome 15 segmental duplications in the human lineage and independent structural changes in apes. The significant clustering (P = 0.002) of breakpoints provides mechanistic evidence for the role of this core duplicon and its palindromic architecture in promoting the evolutionary and disease-related instability of chromosome 15.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Cooper, G.M. et al. A copy number variation morbidity map of developmental delay. Nat. Genet. 43, 838–846 (2011).
Kaminsky, E.B. et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet. Med. 13, 777–784 (2011).
Sharp, A.J. et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 40, 322–328 (2008).
Helbig, I. et al. 15q13.3 microdeletions increase risk of idiopathic generalized epilepsy. Nat. Genet. 41, 160–162 (2009).
International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455, 237–241 (2008).
Stefansson, H. et al. Large recurrent microdeletions associated with schizophrenia. Nature 455, 232–236 (2008).
Miller, D.T. et al. Microdeletion/duplication at 15q13.2q13.3 among individuals with features of autism and other neuropsychiatric disorders. J. Med. Genet. 46, 242–248 (2009).
Shinawi, M. et al. A small recurrent deletion within 15q13.3 is associated with a range of neurodevelopmental phenotypes. Nat. Genet. 41, 1269–1271 (2009).
Williams, N.M. et al. Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: the role of rare variants and duplications at 15q13.3. Am. J. Psychiatry 169, 195–204 (2012).
Kidd, J.M. et al. Mapping and sequencing of structural variation from eight human genomes. Nature 453, 56–64 (2008).
Antonacci, F. et al. Characterization of six human disease-associated inversion polymorphisms. Hum. Mol. Genet. 18, 2555–2566 (2009).
Pujana, M.A. et al. Additional complexity on human chromosome 15q: identification of a set of newly recognized duplicons (LCR15) on 15q11-q13, 15q24, and 15q26. Genome Res. 11, 98–111 (2001).
Pujana, M.A. et al. Human chromosome 15q11-q14 regions of rearrangements contain clusters of LCR15 duplicons. Eur. J. Hum. Genet. 10, 26–35 (2002).
Bailey, J.A. et al. Recent segmental duplications in the human genome. Science 297, 1003–1007 (2002).
Zody, M.C. et al. Analysis of the DNA sequence and duplication history of human chromosome 15. Nature 440, 671–675 (2006).
Jiang, Z. et al. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat. Genet. 39, 1361–1368 (2007).
Sudmant, P.H. et al. Evolution and diversity of copy number variation in the great ape lineage. Genome Res. 23, 1373–1382 (2013).
Huddleston, J. et al. Reconstructing complex regions of genomes using long-read sequencing technology. Genome Res. 24, 688–696 (2014).
Sudmant, P.H. et al. Diversity of human copy number variation and multicopy genes. Science 330, 641–646 (2010).
Dennis, M.Y. et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 149, 912–922 (2012).
Itsara, A. et al. Resolving the breakpoints of the 17q21.31 microdeletion syndrome with next-generation sequencing. Am. J. Hum. Genet. 90, 599–613 (2012).
She, X. et al. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature 431, 927–930 (2004).
Antonacci, F. et al. A large and complex structural polymorphism at 16p12.1 underlies microdeletion disease risk. Nat. Genet. 42, 745–750 (2010).
1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Hardenbol, P. et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 21, 673–678 (2003).
O'Roak, B.J. et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science 338, 1619–1622 (2012).
Zody, M.C. et al. Evolutionary toggling of the MAPT 17q21.31 inversion region. Nat. Genet. 40, 1076–1083 (2008).
Girirajan, S. et al. Refinement and discovery of new hotspots of copy-number variation associated with autism spectrum disorder. Am. J. Hum. Genet. 92, 221–237 (2013).
Steinberg, K.M. et al. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat. Genet. 44, 872–880 (2012).
Sharp, A.J. et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 38, 1038–1042 (2006).
Meyer, M. et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature 505, 403–406 (2014).
Prüfer, K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014).
Mefford, H.C. et al. Further clinical and molecular delineation of the 15q24 microdeletion syndrome. J. Med. Genet. 49, 110–118 (2012).
Wat, M.J. et al. Recurrent microdeletions of 15q25.2 are associated with increased risk of congenital diaphragmatic hernia, cognitive deficits and possibly Diamond-Blackfan anaemia. J. Med. Genet. 47, 777–781 (2010).
Amos-Landgraf, J.M. et al. Chromosome breakage in the Prader-Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am. J. Hum. Genet. 65, 370–386 (1999).
El-Hattab, A.W. et al. Redefined genomic architecture in 15q24 directed by patient deletion/duplication breakpoint mapping. Hum. Genet. 126, 589–602 (2009).
Marques-Bonet, T. et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457, 877–881 (2009).
Locke, D.P. et al. Refinement of a chimpanzee pericentric inversion breakpoint to a segmental duplication cluster. Genome Biol. 4, R50 (2003).
Giannuzzi, G. et al. Hominoid fission of chromosome 14/15 and the role of segmental duplications. Genome Res. 23, 1763–1773 (2013).
Sharp, A.J. et al. Characterization of a recurrent 15q24 microdeletion syndrome. Hum. Mol. Genet. 16, 567–572 (2007).
Lupski, J.R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 14, 417–422 (1998).
Bengesser, K. et al. A novel third type of recurrent NF1 microdeletion mediated by nonallelic homologous recombination between LRRC37B-containing low-copy repeats in 17q11.2. Hum. Mutat. 31, 742–751 (2010).
Gordenin, D.A. et al. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol. Cell. Biol. 13, 5315–5322 (1993).
Leach, D.R. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays 16, 893–900 (1994).
Collick, A. et al. Instability of long inverted repeats within mouse transgenes. EMBO J. 15, 1163–1171 (1996).
Akgün, E. et al. Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol. 17, 5559–5570 (1997).
Ruskin, B. & Fink, G.R. Mutations in POL1 increase the mitotic instability of tandem inverted repeats in Saccharomyces cerevisiae. Genetics 134, 43–56 (1993).
Lemoine, F.J., Degtyareva, N.P., Lobachev, K. & Petes, T.D. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120, 587–598 (2005).
Inagaki, H. et al. Two sequential cleavage reactions on cruciform DNA structures cause palindrome-mediated chromosomal translocations. Nat. Commun. 4, 1592 (2013).
Tanaka, H., Bergstrom, D.A., Yao, M.C. & Tapscott, S.J. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat. Genet. 37, 320–327 (2005).
Carvalho, C.M. et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat. Genet. 43, 1074–1081 (2011).
Lee, J.A., Carvalho, C.M. & Lupski, J.R.A. DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131, 1235–1247 (2007).
Hastings, P.J., Ira, G. & Lupski, J.R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 5, e1000327 (2009).
Payen, C., Koszul, R., Dujon, B. & Fischer, G. Segmental duplications arise from Pol32-dependent repair of broken forks through two alternative replication-based mechanisms. PLoS Genet. 4, e1000175 (2008).
Parsons, J.D. Miropeats: graphical DNA sequence comparisons. Comput. Appl. Biosci. 11, 615–619 (1995).
Smith, J.J., Stuart, A.B., Sauka-Spengler, T., Clifton, S.W. & Amemiya, C.T. Development and analysis of a germline BAC resource for the sea lamprey, a vertebrate that undergoes substantial chromatin diminution. Chromosoma 119, 381–389 (2010).
Alkan, C. et al. Personalized copy number and segmental duplication maps using next-generation sequencing. Nat. Genet. 41, 1061–1067 (2009).
Altschul, S.F., Gish, W., Miller, W., Myers, E.W. & Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Larkin, M.A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007).
Igartua, C. et al. Targeted enrichment of specific regions in the human genome by array hybridization. Curr. Protoc. Hum. Genet. Chapter 18 Unit 18.3 (2010).
Nuttle, X. et al. Rapid and accurate large-scale genotyping of duplicated genes and discovery of interlocus gene conversions. Nat. Methods 10, 903–909 (2013).
Acknowledgements
We thank M. Ventura, C. Campbell and H.C. Mefford for useful discussions and T. Brown for critical review of the manuscript. We also thank S. Diede, H. Tanaka, B. Brewer, C. Payen, L. Harshman and K. Penewit for experimental advice and support for the palindromic snapback assay. We are grateful to all of the families at the participating Simons Simplex Collection (SSC) sites, as well as the principal investigators (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, R. Goin-Kochel, E. Hanson, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, K. Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, Z. Warren and E. Wijsman). We appreciate obtaining access to phenotypic data on SFARI Base. This work was supported, in part, by US National Institutes of Health grants HG002385 and HG004120 to E.E.E. M.Y.D. is supported by the National Institute of Neurological Disorder and Stroke of the US National Institutes of Health (award K99NS083627). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
F.A., M.Y.D. and E.E.E. designed the study. F.A. performed FISH experiments, library construction for Illumina sequencing, array CGH experiments and sequence analysis. M.Y.D. performed MIP experiments, library construction for Illumina sequencing, array CGH experiments and sequence analysis. J.H. performed SMRT sequence analysis and haplotype reconstruction. P.H.S. and K.M.S. performed sequencing data analysis. T.A.G. and R.K.W. performed capillary sequencing and analysis of CH17 and nonhuman primate BAC clones. L.V. and M. Malig performed FISH experiments. M. Miroballo performed array CGH experiments. B.M. performed library construction for SMRT sequencing. L.D. performed MIP experiments and library construction for SMRT sequencing. A.R. performed SMRT sequence analysis. C.T.A., A.S. and J.T. performed library construction for the VMRC53, VMRC54 and VMRC57 BACs. J.A.R. and L.G.S. contributed to 15q13.3 microdeletion data collection. F.A., M.Y.D. and E.E.E. contributed to data interpretation. F.A., M.Y.D. and E.E.E. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc., and was an SAB member of Pacific Biosciences, Inc. (2009–2013) and SynapDx Corp. (2011–2013). J.A.R. is an employee of Signature Genomic Laboratories, a subsidiary of PerkinElmer, Inc. L.G.S. was an employee of Signature Genomic Laboratories and is now an employee of Genetic Veterinary Sciences, Inc.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Figures 1–26 and Supplementary Tables 1, 4, 6–8, 11, 16, 17 and 22–24. (PDF 4075 kb)
Supplementary Tables 2, 3, 5, 9, 10, 12–15 and 18–21
Supplementary Tables 2, 3, 5, 9, 10, 12–15 and 18–21 (XLSX 1291 kb)
Rights and permissions
About this article
Cite this article
Antonacci, F., Dennis, M., Huddleston, J. et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13.3 microdeletion and evolutionary instability. Nat Genet 46, 1293–1302 (2014). https://doi.org/10.1038/ng.3120
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3120
This article is cited by
-
Impaired OTUD7A-dependent Ankyrin regulation mediates neuronal dysfunction in mouse and human models of the 15q13.3 microdeletion syndrome
Molecular Psychiatry (2023)
-
A rare familial rearrangement of chromosomes 9 and 15 associated with intellectual disability: a clinical and molecular study
Molecular Cytogenetics (2021)
-
Selection shapes the landscape of functional variation in wild house mice
BMC Biology (2021)
-
De novo single-nucleotide and copy number variation in discordant monozygotic twins reveals disease-related genes
European Journal of Human Genetics (2019)
-
BAMSI: a multi-cloud service for scalable distributed filtering of massive genome data
BMC Bioinformatics (2018)