Abstract
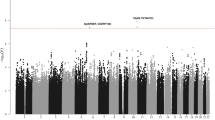
Genome-wide association studies (GWAS) have previously identified 13 loci associated with risk of chronic lymphocytic leukemia or small lymphocytic lymphoma (CLL). To identify additional CLL susceptibility loci, we conducted the largest meta-analysis for CLL thus far, including four GWAS with a total of 3,100 individuals with CLL (cases) and 7,667 controls. In the meta-analysis, we identified ten independent associated SNPs in nine new loci at 10q23.31 (ACTA2 or FAS (ACTA2/FAS), P = 1.22 × 10−14), 18q21.33 (BCL2, P = 7.76 × 10−11), 11p15.5 (C11orf21, P = 2.15 × 10−10), 4q25 (LEF1, P = 4.24 × 10−10), 2q33.1 (CASP10 or CASP8 (CASP10/CASP8), P = 2.50 × 10−9), 9p21.3 (CDKN2B-AS1, P = 1.27 × 10−8), 18q21.32 (PMAIP1, P = 2.51 × 10−8), 15q15.1 (BMF, P = 2.71 × 10−10) and 2p22.2 (QPCT, P = 1.68 × 10−8), as well as an independent signal at an established locus (2q13, ACOXL, P = 2.08 × 10−18). We also found evidence for two additional promising loci below genome-wide significance at 8q22.3 (ODF1, P = 5.40 × 10−8) and 5p15.33 (TERT, P = 1.92 × 10−7). Although further studies are required, the proximity of several of these loci to genes involved in apoptosis suggests a plausible underlying biological mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Albright, F., Teerlink, C., Werner, T.L. & Cannon-Albright, L.A. Significant evidence for a heritable contribution to cancer predisposition: a review of cancer familiality by site. BMC Cancer 12, 138 (2012).
Goldin, L.R., Bjorkholm, M., Kristinsson, S.Y., Turesson, I. & Landgren, O. Elevated risk of chronic lymphocytic leukemia and other indolent non-Hodgkin's lymphomas among relatives of patients with chronic lymphocytic leukemia. Haematologica 94, 647–653 (2009).
Di Bernardo, M.C. et al. A genome-wide association study identifies six susceptibility loci for chronic lymphocytic leukemia. Nat. Genet. 40, 1204–1210 (2008).
Crowther-Swanepoel, D. et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nat. Genet. 42, 132–136 (2010).
Slager, S.L. et al. Genome-wide association study identifies a novel susceptibility locus at 6p21.3 among familial CLL. Blood 117, 1911–1916 (2011).
Slager, S.L. et al. Common variation at 6p21.31 (BAK1) influences the risk of chronic lymphocytic leukemia. Blood 120, 843–846 (2012).
Park, J.H. et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat. Genet. 42, 570–575 (2010).
Wang, Z. et al. Improved imputation of common and uncommon SNPs with a new reference set. Nat. Genet. 44, 6–7 (2012).
Conde, L. et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat. Genet. 42, 661–664 (2010).
1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010).
Howie, B.N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Crowther-Swanepoel, D. et al. Common genetic variation at 15q25.2 impacts on chronic lymphocytic leukaemia risk. Br. J. Haematol. 154, 229–233 (2011).
Rafnar, T. et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 41, 221–227 (2009).
Shete, S. et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat. Genet. 41, 899–904 (2009).
McKay, J.D. et al. Lung cancer susceptibility locus at 5p15.33. Nat. Genet. 40, 1404–1406 (2008).
Wang, Y. et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet. 40, 1407–1409 (2008).
Petersen, G.M. et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat. Genet. 42, 224–228 (2010).
Haiman, C.A. et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor–negative breast cancer. Nat. Genet. 43, 1210–1214 (2011).
Kote-Jarai, Z. et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat. Genet. 43, 785–791 (2011).
Sheng, X. et al. TERT polymorphisms modify the risk of acute lymphoblastic leukemia in Chinese children. Carcinogenesis 34, 228–235 (2013).
Yang, J. et al. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42, 565–569 (2010).
Lee, S.H., Wray, N.R., Goddard, M.E. & Visscher, P.M. Estimating missing heritability for disease from genome-wide association studies. Am. J. Hum. Genet. 88, 294–305 (2011).
Fisher, G.H. et al. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell 81, 935–946 (1995).
Drappa, J., Vaishnaw, A.K., Sullivan, K.E., Chu, J.L. & Elkon, K.B. Fas gene mutations in the Canale-Smith syndrome, an inherited lymphoproliferative disorder associated with autoimmunity. N. Engl. J. Med. 335, 1643–1649 (1996).
Straus, S.E. et al. The development of lymphomas in families with autoimmune lymphoproliferative syndrome with germline Fas mutations and defective lymphocyte apoptosis. Blood 98, 194–200 (2001).
Baseggio, L. et al. In non-follicular lymphoproliferative disorders, IGH/BCL2-fusion is not restricted to chronic lymphocytic leukaemia. Br. J. Haematol. 158, 489–498 (2012).
Cleary, M.L., Smith, S.D. & Sklar, J. Cloning and structural analysis of cDNAs for bcl-2 and a hybrid bcl-2/immunoglobulin transcript resulting from the t(14;18) translocation. Cell 47, 19–28 (1986).
Reshmi, G. et al. C-T variant in a miRNA target site of BCL2 is associated with increased risk of human papilloma virus related cervical cancer—an in silico approach. Genomics 98, 189–193 (2011).
Cheung, V.G. et al. Polymorphic cis- and trans-regulation of human gene expression. PLoS Biol. 8, e1000480 (2010).
Strasser, A., Harris, A.W. & Cory, S. E mu-bcl-2 transgene facilitates spontaneous transformation of early pre-B and immunoglobulin-secreting cells but not T cells. Oncogene 8, 1–9 (1993).
Wensveen, F.M. et al. BH3-only protein Noxa regulates apoptosis in activated B cells and controls high-affinity antibody formation. Blood 119, 1440–1449 (2012).
Smit, L.A. et al. Differential Noxa/Mcl-1 balance in peripheral versus lymph node chronic lymphocytic leukemia cells correlates with survival capacity. Blood 109, 1660–1668 (2007).
Morales, A.A. et al. Expression and transcriptional regulation of functionally distinct Bmf isoforms in B-chronic lymphocytic leukemia cells. Leukemia 18, 41–47 (2004).
Labi, V. et al. Loss of the BH3-only protein Bmf impairs B cell homeostasis and accelerates gamma irradiation–induced thymic lymphoma development. J. Exp. Med. 205, 641–655 (2008).
Crowther-Swanepoel, D. et al. Verification that common variation at 2q37.1, 6p25.3, 11q24.1, 15q23, and 19q13.32 influences chronic lymphocytic leukaemia risk. Br. J. Haematol. 150, 473–479 (2010).
Lan, Q. et al. Genetic susceptibility for chronic lymphocytic leukemia among Chinese in Hong Kong. Eur. J. Haematol. 85, 492–495 (2010).
Nieters, A. et al. PRRC2A and BCL2L11 gene variants influence risk of non-Hodgkin lymphoma: results from the InterLymph consortium. Blood 120, 4645–4648 (2012).
Egle, A., Harris, A.W., Bouillet, P. & Cory, S. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc. Natl. Acad. Sci. USA 101, 6164–6169 (2004).
Kelly, J.L. et al. Germline variation in apoptosis pathway genes and risk of non-Hodgkin's lymphoma. Cancer Epidemiol. Biomarkers Prev. 19, 2847–2858 (2010).
Cox, A. et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet. 39, 352–358 (2007).
Abnet, C.C. et al. Genotypic variants at 2q33 and risk of esophageal squamous cell carcinoma in China: a meta-analysis of genome-wide association studies. Hum. Mol. Genet. 21, 2132–2141 (2012).
Barrett, J.H. et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nat. Genet. 43, 1108–1113 (2011).
Enjuanes, A. et al. Genetic variants in apoptosis and immunoregulation-related genes are associated with risk of chronic lymphocytic leukemia. Cancer Res. 68, 10178–10186 (2008).
Lan, Q. et al. Genetic variants in caspase genes and susceptibility to non-Hodgkin lymphoma. Carcinogenesis 28, 823–827 (2007).
Reya, T. et al. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 423, 409–414 (2003).
Gutierrez, A. Jr. et al. LEF-1 is a prosurvival factor in chronic lymphocytic leukemia and is expressed in the preleukemic state of monoclonal B-cell lymphocytosis. Blood 116, 2975–2983 (2010).
Cunnington, M.S., Santibanez Koref, M., Mayosi, B.M., Burn, J. & Keavney, B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 6, e1000899 (2010).
Koufos, A. et al. Familial Wiedemann-Beckwith syndrome and a second Wilms tumor locus both map to 11p15.5. Am. J. Hum. Genet. 44, 711–719 (1989).
Weksberg, R., Shuman, C. & Beckwith, J.B. Beckwith-Wiedemann syndrome. Eur. J. Hum. Genet. 18, 8–14 (2010).
Morton, L.M. et al. Proposed classification of lymphoid neoplasms for epidemiologic research from the Pathology Working Group of the International Lymphoma Epidemiology Consortium (InterLymph). Blood 110, 695–708 (2007).
Turner, J.J. et al. InterLymph hierarchical classification of lymphoid neoplasms for epidemiologic research based on the WHO classification (2008): update and future directions. Blood 116, e90–e98 (2010).
Pritchard, J.K., Stephens, M. & Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000).
Price, A.L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006).
Ward, L.D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Dixon, A.L. et al. A genome-wide association study of global gene expression. Nat. Genet. 39, 1202–1207 (2007).
Stranger, B.E. et al. Population genomics of human gene expression. Nat. Genet. 39, 1217–1224 (2007).
Pharoah, P.D. et al. Polygenic susceptibility to breast cancer and implications for prevention. Nat. Genet. 31, 33–36 (2002).
Yang, J., Lee, S.H., Goddard, M.E. & Visscher, P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Fearnhead, P. SequenceLDhot: detecting recombination hotspots. Bioinformatics 22, 3061–3066 (2006).
Fearnhead, P., Harding, R.M., Schneider, J.A., Myers, S. & Donnelly, P. Application of coalescent methods to reveal fine-scale rate variation and recombination hotspots. Genetics 167, 2067–2081 (2004).
Li, N. & Stephens, M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165, 2213–2233 (2003).
Crawford, D.C. et al. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat. Genet. 36, 700–706 (2004).
Luna, A. & Nicodemus, K.K. snp.plotter: an R-based SNP/haplotype association and linkage disequilibrium plotting package. Bioinformatics 23, 774–776 (2007).
Acknowledgements
We thank C. Allmer, E. Angelucci, A. Bigelow, I. Brock, K. Butterbach, A. Chabrier, D. Chan-Lam, J.M. Conners, D. Connley, M. Cornelis, K. Corsano, C. Dalley, D. Cox, H. Cramp, R. Cutting, H. Dykes, L. Ershler, A. Gabbas, R.P. Gallagher, R.D. Gascoyne, P. Hui, L. Irish, L. Jacobus, S. Kaul, J. Lunde, M. McAdams, R. Montalvan, M. Rais, T. Rattle, L. Rigacci, K. Snyder, G. Specchia, M. Stagner, P. Taylor, G. Thomas, C. Tornow, G. Wood, M. Yang and M. Zucca for their assistance. The overall GWAS project was supported by the intramural program of the Division of Cancer Epidemiology and Genetics, NCI, US National Institutes of Health. A full list of acknowledgments is provided in the Supplementary Note.
Author information
Authors and Affiliations
Contributions
S.I.B., C.F.S., N.J.C., A.N., W.C., S.S.W., L.R.T., A.R.B.-W., P.H., M.P.P., B.M.B., B.K.A., P.C., Y.Z., G.S., A.Z.-J., C.L., K.E.S., J.M., P.V., J.J.S., A.K., S.d.S., H.H., J.R.C., S.J.C., N.R. and S.L.S. organized and designed the study. C.F.S., N.J.C., B.J., L.B., J.Y., A.H., L.C., P.M.B., E.A.H., J.M.C., J.R.C., S.J.C. and S.L.S. conducted and supervised the genotyping of samples. S.I.B., C.F.S., V.J., N.J.C., Z.W., N.C., C.C.C., M.Y., K.B.J., L.L., J.S., J.-H.P., J.R.C., L.C., S.J.C., N.R. and S.L.S. contributed to the design and execution of statistical analyses. S.I.B., C.F.S., V.J., N.J.C., A.N., Z.W., W.C., A.M., R.S.K., N.C., C.C.C., M.Y., C.L., H.H., J.R.C., S.J.C., N.R. and S.L.S. wrote the first draft of the manuscript. S.I.B., C.F.S., V.J., N.J.C., A.N., W.C., A.M., S.S.W., R.S.K., Q.L., L.R.T., A.R.B.-W., P.H., M.P.P., B.M.B., B.K.A., P.C., Y.Z., G.S., A.Z.-J., T.G.C., T.D.S., A.J.N., N.E.K., M.L., A.H.W., K.E.S., H.-O.A., M. Melbye, B.G., E.T.C., M.G., K.C., L.A.C.-A., B.J., W.R.D., B.K.L., G.J.W., L.C., P.M.B., J.R., E.A.H., M.T.S., R.D.J., L.F.T., S.d.S., Y.B., N.B., P. Boffetta, P. Brennan, L.F., M. Maynadie, J.M., A.S., K.G.R., S.J.A., C.M. Vachon, L.R.G., S.S.S., M.C.L., L.G.S., J.F.L., J.M.C., J.B.W., V.A.M., N.E.C., A.D.N., M.S.L., A.J.D.R., L.M.M., R.K.S., E.R., P.V., R.K., D.T., G.M., E.W., M.-D.C., R.C.H.V., R.C.T., G.G.G., D.A., J.V., S.W., J.C., T.Z., T.R.H., K.O., A.Z., R.J.K., J.J.S., K.A.B., F.L., E.G., P.K., A.K., J.T., C.M. Vajdic, M.G.E., G.M.F., L.M., L.L., J.A.S., S.C., J.F.F., K.E.N., A. Cox, J.S., J.W., A. Carracedo, C.L.-O., S.B., I.S., D.M.-G., E.C., H.H., J.R.C., N.R. and S.L.S. conducted the epidemiological studies and contributed samples to the GWAS and/or follow-up genotyping. All authors contributed to the writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–9, Supplementary Figures 1–4 and Supplementary Note (PDF 917 kb)
Rights and permissions
About this article
Cite this article
Berndt, S., Skibola, C., Joseph, V. et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet 45, 868–876 (2013). https://doi.org/10.1038/ng.2652
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2652
This article is cited by
-
Multi-omics analysis identifies RFX7 targets involved in tumor suppression and neuronal processes
Cell Death Discovery (2023)
-
Implementation of individualised polygenic risk score analysis: a test case of a family of four
BMC Medical Genomics (2022)
-
Distinct germline genetic susceptibility profiles identified for common non-Hodgkin lymphoma subtypes
Leukemia (2022)
-
Ethnic and geographic diversity of chronic lymphocytic leukaemia
Leukemia (2021)
-
Genome-wide association study identifies risk loci for progressive chronic lymphocytic leukemia
Nature Communications (2021)