Abstract
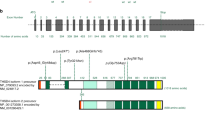
A predisposition for thoracic aortic aneurysms leading to acute aortic dissections can be inherited in families in an autosomal dominant manner. Genome-wide linkage analysis of two large unrelated families with thoracic aortic disease followed by whole-exome sequencing of affected relatives identified causative mutations in TGFB2. These mutations—a frameshift mutation in exon 6 and a nonsense mutation in exon 4—segregated with disease with a combined logarithm of odds (LOD) score of 7.7. Sanger sequencing of 276 probands from families with inherited thoracic aortic disease identified 2 additional TGFB2 mutations. TGFB2 encodes transforming growth factor (TGF)-β2, and the mutations are predicted to cause haploinsufficiency for TGFB2; however, aortic tissue from cases paradoxically shows increased TGF-β2 expression and immunostaining. Thus, haploinsufficiency for TGFB2 predisposes to thoracic aortic disease, suggesting that the initial pathway driving disease is decreased cellular TGF-β2 levels leading to a secondary increase in TGF-β2 production in the diseased aorta.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
References
Hiratzka, L.F. et al. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with thoracic aortic disease: executive summary. A report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines, American Association for Thoracic Surgery, American College of Radiology, American Stroke Association, Society of Cardiovascular Anesthesiologists, Society for Cardiovascular Angiography and Interventions, Society of Interventional Radiology, Society of Thoracic Surgeons, and Society for Vascular Medicine. Circulation 121, e266–e369 (2010).
Faivre, L. et al. Pathogenic FBN1 mutations in 146 adults not meeting clinical diagnostic criteria for Marfan syndrome: further delineation of type 1 fibrillinopathies and focus on patients with an isolated major criterion. Am. J. Med. Genet. A. 149A, 854–860 (2009).
Loeys, B.L. et al. Aneurysm syndromes caused by mutations in the TGF-β receptor. N. Engl. J. Med. 355, 788–798 (2006).
van de Laar, I.M. et al. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat. Genet. 43, 121–126 (2011).
Gomez, D. et al. Syndromic and non-syndromic aneurysms of the human ascending aorta share activation of the Smad2 pathway. J. Pathol. 218, 131–142 (2009).
Habashi, J.P. et al. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science 312, 117–121 (2006).
Regalado, E. et al. Autosomal dominant inheritance of a predisposition to thoracic aortic aneurysms and dissections and intracranial saccular aneurysms. Am. J. Med. Genet. A. 155A, 2125–2130 (2011).
Prakash, S.K. et al. Rare copy number variants disrupt genes regulating vascular smooth muscle cell adhesion and contractility in sporadic thoracic aortic aneurysms and dissections. Am. J. Hum. Genet. 87, 743–756 (2010).
Silva, A.L. & Romao, L. The mammalian nonsense-mediated mRNA decay pathway: to decay or not to decay! Which players make the decision? FEBS Lett. 583, 499–505 (2009).
Loeys, B.L. et al. The revised Ghent nosology for the Marfan syndrome. J. Med. Genet. 47, 476–485 (2010).
Attias, D. et al. Comparison of clinical presentations and outcomes between patients with TGFBR2 and FBN1 mutations in Marfan syndrome and related disorders. Circulation 120, 2541–2549 (2009).
Sanford, L.P. et al. TGFβ2 knockout mice have multiple developmental defects that are non-overlapping with other TGFβ knockout phenotypes. Development 124, 2659–2670 (1997).
Majesky, M.W. Developmental basis of vascular smooth muscle diversity. Arterioscler. Thromb. Vasc. Biol. 27, 1248–1258 (2007).
Chen, S. & Lechleider, R.J. Transforming growth factor-β–induced differentiation of smooth muscle from a neural crest stem cell line. Circ. Res. 94, 1195–1202 (2004).
Inamoto, S. et al. TGFBR2 mutations alter smooth muscle cell phenotype and predispose to thoracic aortic aneurysms and dissections. Cardiovasc. Res. 88, 520–529 (2010).
Regalado, E.S. et al. Exome sequencing identifies SMAD3 mutations as a cause of familial thoracic aortic aneurysm and dissection with intracranial and other arterial aneurysms. Circ. Res. 109, 680–686 (2011).
van de Laar, I.M. et al. Phenotypic spectrum of the SMAD3-related aneurysms-osteoarthritis syndrome. J. Med. Genet. 49, 47–57 (2012).
Mizuguchi, T. et al. Heterozygous TGFBR2 mutations in Marfan syndrome. Nat. Genet. 36, 855–860 (2004).
Stheneur, C. et al. Identification of 23 TGFBR2 and 6 TGFBR1 gene mutations and genotype-phenotype investigations in 457 patients with Marfan syndrome type I and II, Loeys-Dietz syndrome and related disorders. Hum. Mutat. 29, E284–E295 (2008).
Gomez, D. et al. Epigenetic control of vascular smooth muscle cells in Marfan and non-Marfan thoracic aortic aneurysms. Cardiovasc. Res. 89, 446–456 (2011).
Iwata, J. et al. Modulation of noncanonical TGF-β signaling prevents cleft palate in Tgfbr2 mutant mice. J. Clin. Invest. 122, 873–885 (2012).
Faivre, L. et al. Effect of mutation type and location on clinical outcome in 1,013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: an international study. Am. J. Hum. Genet. 81, 454–466 (2007).
Cottingham, R.W. Jr., Idury, R.M. & Schaffer, A.A. Faster sequential genetic linkage computations. Am. J. Hum. Genet. 53, 252–263 (1993).
Kong, A. et al. A high-resolution recombination map of the human genome. Nat. Genet. 31, 241–247 (2002).
O'Connell, J.R. & Weeks, D.E. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 63, 259–266 (1998).
Fishelson, M. & Geiger, D. Exact genetic linkage computations for general pedigrees. Bioinformatics 18 (suppl. 1), S189–S198 (2002).
Weeks, D.E., Sobel, E., O'Connell, J.R. & Lange, K. Computer programs for multilocus haplotyping of general pedigrees. Am. J. Hum. Genet. 56, 1506–1507 (1995).
Lindner, T.H. & Hoffmann, K. easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics 21, 405–407 (2005).
O'Roak, B.J. et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat. Genet. 43, 585–589 (2011).
Ng, S.B. et al. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 42, 30–35 (2010).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
DePristo, M.A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011).
He, R. et al. Characterization of the inflammatory and apoptotic cells in the aortas of patients with ascending thoracic aortic aneurysms and dissections. J. Thorac. Cardiovasc. Surg. 131, 671–678 (2006).
Guo, D.C. & Milewicz, D.M. Methodology for using a universal primer to label amplified DNA segments for molecular analysis. Biotechnol. Lett. 25, 2079–2083 (2003).
Acknowledgements
The authors are extremely grateful to the families involved in this study and to the medical professionals who aided in the collection of clinical data from the families. We would like to thank the NHLBI GO Exome Sequencing Project and its ongoing studies that produced and provided exome variant calls for comparison: the Lung Cohorts Sequencing Project (HL-102923), the NHLBI Women's Health Initiative Sequencing Project (HL-102924), the Heart Cohorts Sequencing Project (HL-103010), the Broad Institute Sequencing Project (HL-102925), the Northwest Genomics Center Sequencing Project (HL-102926 to D.A.N., M.J.R. and J.S.) and the Family Studies Project Team. We thank J.M. Serfaty for providing vascular imaging and L. Louedec and Z. Ren for technical assistance.
The following sources provided funding for these studies: the US National Institutes of Health (NIH) RO1 HL62594 (D.M.M.), NIH P50HL083794-01 (D.M.M.), NIH UL1 RR024148 (Centers for Clinical and Translational Sciences), the Vivian L. Smith Foundation (D.M.M.), the TexGen Foundation (D.M.M.), Richard T. Pasani Funds (D.M.M.), Groupement d'Interet Scientifique (GIS)–Maladies Rares (C.B.), Projet Hospitalier de Recherche Clinique (PHRC) AOM09093 (G.J.), PHRC AOM 10108 (C.B.), Agence Nationale de Recherche (ANR) 2010 BLAN 1129 from the French National Research Agency (G.J.) and the European Union Framework Programme 7–integrated project Fighting Aneurysmal Disease (FAD, see URLs) (J.B.M.).
Author information
Authors and Affiliations
Consortia
Contributions
C.B., D.M.M. and G.J. planned the project, designed the experiments, oversaw all aspects of the research and wrote the manuscript. C.B., S.M.L., R.L.P.S.-C., M.V., D.-C.G., M.-S.G. and M.A. performed linkage analyses and analyzed exome sequencing data. M.J.R., J.S. and D.A.N. performed the exome sequencing in TAA288. D.-C.G., A.H.L. and N.H. analyzed exome sequencing data and performed Sanger sequencing analysis with H.d'I., L.G., D.-C.G. and C.S.K. performed the TGFB2 transcript and protein assays. J.B.M. performed histological and imunostaining analyses with M.-S.G. S.K.P. carried out the genomic deletion analysis. E.S.R., B.G., C.M., A.C.B. and L. Gouya provided clinical and pedigree data. D.D., D.M.M. and G.J. provided clinical data and performed sample collection. A.V. revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
A list of members and affiliations is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Tables 1–3 and Supplementary Figures 1–3 (PDF 387 kb)
Rights and permissions
About this article
Cite this article
Boileau, C., Guo, DC., Hanna, N. et al. TGFB2 mutations cause familial thoracic aortic aneurysms and dissections associated with mild systemic features of Marfan syndrome. Nat Genet 44, 916–921 (2012). https://doi.org/10.1038/ng.2348
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.2348
This article is cited by
-
Diagnosis and treatment of cardiovascular disease in patients with heritable connective tissue disorders or heritable thoracic aortic diseases
Cardiovascular Intervention and Therapeutics (2024)
-
Genome-wide association study of varicose veins identifies a protective missense variant in GJD3 enriched in the Finnish population
Communications Biology (2023)
-
Genetics and mechanisms of thoracic aortic disease
Nature Reviews Cardiology (2023)
-
Aortic aneurysms: current pathogenesis and therapeutic targets
Experimental & Molecular Medicine (2023)
-
Incorporation of DNA methylation quantitative trait loci (mQTLs) in epigenome-wide association analysis: application to birthweight effects in neonatal whole blood
Clinical Epigenetics (2022)