Abstract
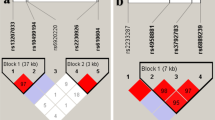
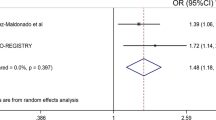
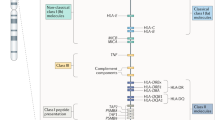
Ankylosing spondylitis is a common form of inflammatory arthritis predominantly affecting the spine and pelvis that occurs in approximately 5 out of 1,000 adults of European descent. Here we report the identification of three variants in the RUNX3, LTBR-TNFRSF1A and IL12B regions convincingly associated with ankylosing spondylitis (P < 5 × 10−8 in the combined discovery and replication datasets) and a further four loci at PTGER4, TBKBP1, ANTXR2 and CARD9 that show strong association across all our datasets (P < 5 × 10−6 overall, with support in each of the three datasets studied). We also show that polymorphisms of ERAP1, which encodes an endoplasmic reticulum aminopeptidase involved in peptide trimming before HLA class I presentation, only affect ankylosing spondylitis risk in HLA-B27–positive individuals. These findings provide strong evidence that HLA-B27 operates in ankylosing spondylitis through a mechanism involving aberrant processing of antigenic peptides.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
11 August 2011
In the version of this article initially published, the name of author Udo Oppermann was incorrectly spelled as Udo Opperman, and the name of author Loukas Moutsianas was incorrectly spelled as Loukas Moutsianis. The errors have been corrected in the HTML and PDF versions of the article.
References
Pedersen, O.B. et al. Ankylosing spondylitis in Danish and Norwegian twins: occurrence and the relative importance of genetic vs. environmental effectors in disease causation. Scand. J. Rheumatol. 37, 120–126 (2008).
Brown, M.A. et al. Susceptibility to ankylosing spondylitis in twins: the role of genes, HLA, and the environment. Arthritis Rheum. 40, 1823–1828 (1997).
Brown, M.A., Laval, S.H., Brophy, S. & Calin, A. Recurrence risk modelling of the genetic susceptibility to ankylosing spondylitis. Ann. Rheum. Dis. 59, 883–886 (2000).
Calin, A., Marder, A., Becks, E. & Burns, T. Genetic differences between B27 positive patients with ankylosing spondylitis and B27 positive healthy controls. Arthritis Rheum. 26, 1460–1464 (1983).
van der Linden, S., Valkenburg, H. & Cats, A. The risk of developing ankylosing spondylitis in HLA-B27 positive individuals: a family and population study. Br. J. Rheumatol. 22, 18–19 (1983).
Reveille, J.D. et al. Genome-wide association study of ankylosing spondylitis identifies non-MHC susceptibility loci. Nat. Genet. 42, 123–127 (2010).
Wellcome Trust Case Control Consortium et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 39, 1329–1337 (2007).
Danoy, P. et al. Association of variants at 1q32 and STAT3 with ankylosing spondylitis suggests genetic overlap with Crohn's disease. PLoS Genet. 6, e1001195 (2010).
van der Linden, S., Valkenburg, H.A. & Cats, A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum. 27, 361–368 (1984).
Pointon, J.J. et al. Elucidating the chromosome 9 association with AS; CARD9 is a candidate gene. Genes and Immunity 12, 319–320 (2011).
Park, J.H. et al. Signaling by intrathymic cytokines, not T cell antigen receptors, specifies CD8 lineage choice and promotes the differentiation of cytotoxic-lineage T cells. Nat. Immunol. 11, 257–264 (2010).
Ferreira, M.A. et al. Quantitative trait loci for CD4:CD8 lymphocyte ratio are associated with risk of type 1 diabetes and HIV-1 immune control. Am. J. Hum. Genet. 86, 88–92 (2010).
Su, Z. et al. Bayesian method for detecting and characterizing allelic heterogeneity and boosting signals in genome-wide association studies. Stat. Sci. 24, 430–450 (2009).
Stephens, M. & Balding, D.J. Bayesian statistical methods for genetic association studies. Nat. Rev. Genet. 10, 681–690 (2009).
Amroun, H. et al. Early-onset ankylosing spondylitis is associated with a functional MICA polymorphism. Hum. Immunol. 66, 1057–1061 (2005).
Yabuki, K. et al. Triplet repeat polymorphism in the MICA gene in HLA-B27 positive and negative Caucasian patients with ankylosing spondylitis. Hum. Immunol. 60, 83–86 (1999).
Brown, M.A. et al. HLA class I associations of ankylosing spondylitis in the white population in the United Kingdom. Ann. Rheum. Dis. 55, 268–270 (1996).
D'Amato, M. et al. Relevance of residue 116 of HLA-B27 in determining susceptibility to ankylosing spondylitis. Eur. J. Immunol. 25, 3199–3201 (1995).
López-Larrea, C. et al. HLA-B27 subtypes in Asian patients with ankylosing spondylitis. Evidence for new associations. Tissue Antigens 45, 169–176 (1995).
Brown, M.A. et al. HLA Class I and II associations of ankylosing spondylitis. Arthritis Rheum. 60, S437 (2009).
Saric, T. et al. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat. Immunol. 3, 1169–1176 (2002).
Chang, S.C., Momburg, F., Bhutani, N. & Goldberg, A.L. The ER aminopeptidase, ERAP1, trims precursors to lengths of MHC class I peptides by a “molecular ruler” mechanism. Proc. Natl. Acad. Sci. USA 102, 17107–17112 (2005).
Cui, X., Rouhani, F.N., Hawari, F. & Levine, S.J. Shedding of the type II IL-1 decoy receptor requires a multifunctional aminopeptidase, aminopeptidase regulator of TNF receptor type 1 shedding. J. Immunol. 171, 6814–6819 (2003).
Cui, X., Rouhani, F.N., Hawari, F. & Levine, S.J. An aminopeptidase, ARTS-1, is required for interleukin-6 receptor shedding. J. Biol. Chem. 278, 28677–28685 (2003).
Cui, X. et al. Identification of ARTS-1 as a novel TNFR1-binding protein that promotes TNFR1 ectodomain shedding. J. Clin. Invest. 110, 515–526 (2002).
Evnouchidou, I., Berardi, M.J. & Stratikos, E. A continuous fluorigenic assay for the measurement of the activity of endoplasmic reticulum aminopeptidase 1: competition kinetics as a tool for enzyme specificity investigation. Anal. Biochem. 395, 33–40 (2009).
Libioulle, C. et al. Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 3, e58 (2007).
Cho, J.H. The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 8, 458–466 (2008).
Zhang, X.J. et al. Psoriasis genome-wide association study identifies susceptibility variants within LCE gene cluster at 1q21. Nat. Genet. 41, 205–210 (2009).
Cargill, M. et al. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am. J. Hum. Genet. 80, 273–290 (2007).
Duerr, R.H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006).
Shen, H., Goodall, J.C. & Hill Gaston, J.S. Frequency and phenotype of peripheral blood Th17 cells in ankylosing spondylitis and rheumatoid arthritis. Arthritis Rheum. 60, 1647–1656 (2009).
Gagliardi, M.C. et al. Endogenous PGE2 promotes the induction of human Th17 responses by fungal β-glucan. J. Leukoc. Biol. 88, 947–954 (2010).
Ruutu, M. et al. Fungal β-glucan triggers spondyloarthropathy and Crohn's disease in SKG mice. Arthritis Rheum. S1, 1446 (2010).
Hirota, K. et al. T cell self-reactivity forms a cytokine milieu for spontaneous development of IL-17+ Th cells that cause autoimmune arthritis. J. Exp. Med. 204, 41–47 (2007).
Armaka, M. et al. Mesenchymal cell targeting by TNF as a common pathogenic principle in chronic inflammatory joint and intestinal diseases. J. Exp. Med. 205, 331–337 (2008).
Risch, N. Assessing the role of HLA-linked and unlinked determinants of disease. Am. J. Hum. Genet. 40, 1–14 (1987).
Risch, N. Linkage strategies for genetically complex traits. I. Multilocus models. Am. J. Hum. Genet. 46, 222–228 (1990).
Strange, A. et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat. Genet. 42, 985–990 (2010).
Jaakkola, E. et al. Finnish HLA studies confirm the increased risk conferred by HLA-B27 homozygosity in ankylosing spondylitis. Ann. Rheum. Dis. 65, 775–780 (2006).
Rudwaleit, M. et al. Effectiveness, safety, and predictors of good clinical response in 1250 patients treated with adalimumab for active ankylosing spondylitis. J. Rheumatol. 36, 801–808 (2009).
Pointon, J.J. et al. The chromosome 16q region associated with ankylosing spondylitis includes the candidate gene tumour necrosis factor receptor type 1-associated death domain (TRADD). Ann. Rheum. Dis. 69, 1243–1246 (2010).
Conrad, D.F. et al. Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712 (2010).
Teo, Y.Y. et al. A genotype calling algorithm for the Illumina BeadArray platform. Bioinformatics 23, 2741–2746 (2007).
Spencer, C.C. et al. Dissection of the genetics of Parkinson's disease identifies an additional association 5′ of SNCA and multiple associated haplotypes at 17q21. Hum. Mol. Genet. 20, 345–353 (2011).
Howie, B.N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Li, Y., Willer, C., Sanna, S. & Abecasis, G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 10, 387–406 (2009).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
R Core Development Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, 2009).
Du, P., Kibbe, W.A. & Lin, S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548 (2008).
Simon, R. et al. Analysis of gene expression data using BRB-array tools. Cancer Inform. 3, 11–17 (2007).
Acknowledgements
We would like to thank all participating subjects with ankylosing spondylitis and healthy individuals who provided the DNA and clinical information necessary for this study. The Wellcome Trust Case Control Consortium 2 project is funded by the Wellcome Trust (083948/Z/07/Z). We also thank S. Bertrand, J. Bryant, S.L. Clark, J.S. Conquer, T. Dibling, J.C. Eldred, S. Gamble, C. Hind, A. Wilk, C.R. Stribling and S. Taylor of the Wellcome Trust Sanger Institute's Sample and Genotyping Facilities for technical assistance. The TASC study was funded by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) grants P01-052915 and R01-AR046208. Funding was also received from the University of Texas at Houston Clinical and Translational Science Awards grant UL1RR024188, Cedars-Sinai General Clinical Research Centre grant MO1-RR00425, Intramural Research Program, NIAMS/US National Institutes of Health and Rebecca Cooper Foundation (Australia). This study was funded, in part, by Arthritis Research UK (Grants 19536 and 18797), by the Wellcome Trust (grant number 076113) and by the Oxford Comprehensive Biomedical Research Centre ankylosing spondylitis chronic disease cohort (theme code: A91202). The Spondyloarthritis Research Consortium of Canada (SPARCC) was funded by a National Research Initiative Award from the Arthritis Society (Canada). G.P.T. was funded by a Lions Medical Research Foundation fellowship. M.A.B. is funded by a National Health and Medical Research Council (Australia) Principal Research Fellowship, and support for this study was received from a National Health and Medical Research Council (Australia) program grant (566938) and project grant (569829) and from the Australian Cancer Research Foundation and Rebecca Cooper Medical Research Foundation. P. Donnelly was supported in part by a Wolfson-Royal Society Merit Award. We are also very grateful for the invaluable support received from the National Ankylosing Spondylitis Society (UK) and Spondyloarthritis Association of America in case recruitment. Additional financial and technical support for subject recruitment was provided by the National Institute for Health Research (NIHR), Oxford Musculoskeletal Biomedical Research Unit and NIHR Thames Valley Comprehensive Local Research Network. We acknowledge use of the British 1958 Birth Cohort DNA collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02, and we thank W. Bodmer and B. Winney for use of the People of the British Isles DNA collection, which was funded by the Wellcome Trust. We would like to thank A. Mathieu (Cagliari University) and S. Brown (Cedars-Sinai Hospital) for providing samples. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust.
Author information
Authors and Affiliations
Consortia
Contributions
M.A.B., L.A.B., C. Farrar, J.D.R., J.J.P., B.P.W., D.G., W.M. and P.R. oversaw cohort collection for the discovery and replication datasets. The WTCCC2 DNA, genotyping, data quality control and informatics group (S.J.B., S.D., S.E., E.G., C.L. and L.P.) executed GWAS sample handling, genotyping and quality control. The WTCCC2 data and analysis group (A.S., C.C.A.S., G.B., C.B., C. Freeman and P. Donnelly), D.M.E. and M.A.B. performed statistical analyses. M.A.B., D.M.E., C.C.A.S. and P. Donnelly contributed to writing the manuscript. The WTCCC2 management committee (J.M.B., E.B., M.A.B., J.P.C., A.C., P. Deloukas, P. Donnelly (chairperson), A. Duncanson, J.J., J.L., H.S.M., C.G.M., C.N.A.P., L.P., R.P., A.R., S.J.S., R.C.T., A.C.V. and N.W.W.) monitored the execution of the GWAS. D.H., G.K. and U.O. performed analyses of recombinant ERAP1 function. T.J.K. and G.P.T. performed gene expression, cell count and ERAP1 sheddase functional studies. Other authors contributed variously to sample collection and all other aspects of the study. All authors reviewed the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The University of Queensland has applied for patents relating to material presented in this manuscript in Australia, Europe and the United States.
Additional information
A full list of members is provided in the Supplementary Note.
A full list of members is provided in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Note, Supplementary Tables 1–9 and Supplementary Figures 1–6. (PDF 1283 kb)
Rights and permissions
About this article
Cite this article
The Australo-Anglo-American Spondyloarthritis Consortium (TASC)., the Wellcome Trust Case Control Consortium 2 (WTCCC2)., Evans, D. et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet 43, 761–767 (2011). https://doi.org/10.1038/ng.873
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.873
This article is cited by
-
Zur Rolle von HLA-B27 in der Pathogenese und Diagnostik der axialen Spondyloarthritis
Zeitschrift für Rheumatologie (2024)
-
Synergistic effects of BTN3A1, SHP2, CD274, and STAT3 gene polymorphisms on the risk of systemic lupus erythematosus: a multifactorial dimensional reduction analysis
Clinical Rheumatology (2024)
-
MICA and NLRP3 gene polymorphisms interact synergistically affecting the risk of ankylosing spondylitis
Immunologic Research (2024)
-
Identification of potential biomarkers for ankylosing spondylitis based on bioinformatics analysis
BMC Musculoskeletal Disorders (2023)
-
IgA nephropathy
Nature Reviews Disease Primers (2023)