Abstract
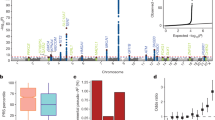
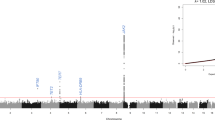
Genome-wide association studies have identified a number of new disease susceptibility loci that represent haplotypes defined by numerous SNPs. SNPs within a disease-associated haplotype are thought to influence either the expression of genes or the sequence of the proteins they encode. In a series of investigations of the JAK2 gene in myeloproliferative neoplasms, we uncovered a new property of haplotypes that can explain their disease association. We observed a nonrandom distribution of the somatic JAK2V617F oncogenic mutation between two parental alleles of the JAK2 gene. We identified a haplotype that preferentially acquires JAK2V617F and confers susceptibility to myeloproliferative neoplasms. One interpretation of our results is that a certain combination of SNPs may render haplotypes differentially susceptible to somatic mutagenesis. Thus, disease susceptibility loci may harbor somatic mutations that have a role in disease pathogenesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
James, C. et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 434, 1144–1148 (2005).
Kralovics, R. et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N. Engl. J. Med. 352, 1779–1790 (2005).
Levine, R.L. et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 7, 387–397 (2005).
Baxter, E.J. et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 365, 1054–1061 (2005).
Scott, L.M. et al. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N. Engl. J. Med. 356, 459–468 (2007).
Pikman, Y. et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 3, e270 (2006).
Pardanani, A.D. et al. MPL515 mutations in myeloproliferative and other myeloid disorders: a study of 1182 patients. Blood 108, 3472–3476 (2006).
Plo, I. et al. JAK2 stimulates homologous recombination and genetic instability: potential implication in the heterogeneity of myeloproliferative disorders. Blood 112, 1402–1412 (2008).
Kralovics, R. et al. Acquisition of the V617F mutation of JAK2 is a late genetic event in a subset of patients with myeloproliferative disorders. Blood 108, 1377–1380 (2006).
Kralovics, R. Genetic complexity of myeloproliferative neoplasms. Leukemia 22, 1841–1848 (2008).
Levi, S. et al. Multiple K-ras codon 12 mutations in cholangiocarcinomas demonstrated with a sensitive polymerase chain reaction technique. Cancer Res. 51, 3497–3502 (1991).
Sozzi, G. et al. Genetic evidence for an independent origin of multiple preneoplastic and neoplastic lung lesions. Cancer Res. 55, 135–140 (1995).
Moskaluk, C.A., Hruban, R.H. & Kern, S.E. p16 and K-ras gene mutations in the intraductal precursors of human pancreatic adenocarcinoma. Cancer Res. 57, 2140–2143 (1997).
Laghi, L. et al. Lack of mutation at codon 531 of SRC in advanced colorectal cancers from Italian patients. Br. J. Cancer 84, 196–198 (2001).
Agaimy, A. et al. Multiple sporadic gastrointestinal stromal tumors (GISTs) of the proximal stomach are caused by different somatic KIT mutations suggesting a field effect. Am. J. Surg. Pathol. 32, 1553–1559 (2008).
Li, S. et al. Clonal heterogeneity in polycythemia vera patients with JAK2 exon12 and JAK2–V617F mutations. Blood 111, 3863–3866 (2008).
Pardanani, A., Fridley, B.L., Lasho, T.L., Gilliland, D.G. & Tefferi, A. Host genetic variation contributes to phenotypic diversity in myeloproliferative disorders. Blood 111, 2785–2789 (2008).
Laken, S.J. et al. Familial colorectal cancer in Ashkenazim due to a hypermutable tract in APC. Nat. Genet. 17, 79–83 (1997).
Mechanic, L.E. et al. Common genetic variation in TP53 is associated with lung cancer risk and prognosis in African Americans and somatic mutations in lung tumors. Cancer Epidemiol. Biomarkers Prev. 16, 214–222 (2007).
Lin, M. et al. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics 20, 1233–1240 (2004).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Barrett, J.C., Fry, B., Maller, J. & Daly, M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265 (2005).
Acknowledgements
The study was supported by funding from the Austrian Academy of Sciences, Austrian Science Fund (FWF, P20033-B11) and the Initiative for Cancer Research of the Medical University of Vienna. We thank C. Ay and N. Bachhofner for help with sample collection and T. Burkard for advice on statistical analysis. We thank H. Pickersgill for help with the manuscript.
Author information
Authors and Affiliations
Contributions
R.K. designed the study and drafted the paper with assistance of D.O. and A.H.; D.O., T.B. and R.J. performed the experiments; D.O. and A.H. performed statistical analyses; T.B., B.G., H.G. and I.P. coordinated and performed the case and control sample collection and clinical management of cases.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1 and 2 (PDF 272 kb)
Rights and permissions
About this article
Cite this article
Olcaydu, D., Harutyunyan, A., Jäger, R. et al. A common JAK2 haplotype confers susceptibility to myeloproliferative neoplasms. Nat Genet 41, 450–454 (2009). https://doi.org/10.1038/ng.341
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.341
This article is cited by
-
The genomic analysis brings a new piece to the molecular jigsaw of idiopathic erythrocytosis
Experimental Hematology & Oncology (2022)
-
CHST15 gene germline mutation is associated with the development of familial myeloproliferative neoplasms and higher transformation risk
Cell Death & Disease (2022)
-
Sulindac acetohydrazide derivative attenuates against cisplatin induced organ damage by modulation of antioxidant and inflammatory signaling pathways
Scientific Reports (2022)
-
Essential thrombocythaemia progression to the fibrotic phase is associated with a decrease in JAK2 and PDL1 levels
Annals of Hematology (2022)
-
Molecular Pathogenesis of Myeloproliferative Neoplasms
Current Hematologic Malignancy Reports (2022)