
Coffea arabica diversification history
Genome assemblies of allotetraploid Coffea arabica and representatives of its diploid progenitors provide insights into diversification history.

Genome assemblies of allotetraploid Coffea arabica and representatives of its diploid progenitors provide insights into diversification history.

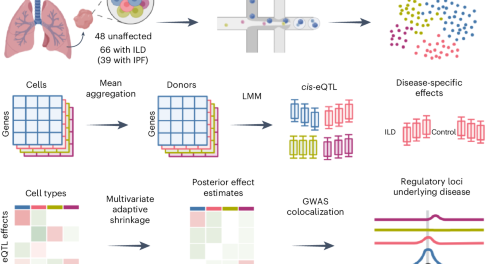
Using single-cell RNA-sequencing (scRNA-seq) of lung tissue, expression quantitative trait loci (eQTLs) were mapped across 38 cell types, revealing both shared and cell-type-specific effects. Highly cell-type-specific disease-interaction eQTLs were linked to cellular dysregulation in lung disease and lung disease risk variants were connected to their regulatory targets in relevant cell types.