
Coffea arabica diversification history
Genome assemblies of allotetraploid Coffea arabica and representatives of its diploid progenitors provide insights into diversification history.

Genome assemblies of allotetraploid Coffea arabica and representatives of its diploid progenitors provide insights into diversification history.

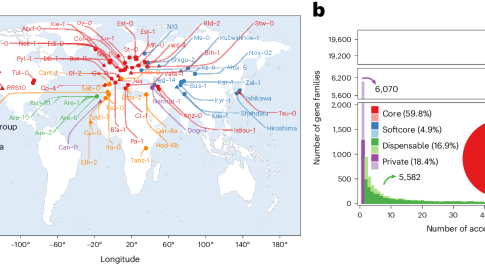
Chromosome-level genome sequences of 69 diverse Arabidopsis thaliana strains reveal a quasi-fixed genome structure worldwide, in which large rearrangement is limited almost exclusively to the centromeric regions. Pan-genome analysis uncovered substantial diversity in gene content that, together with the genome assemblies, will fuel future genetic research.