Abstract
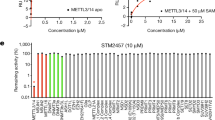
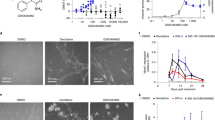
Protein arginine methyltransferase-5 (PRMT5) is reported to have a role in diverse cellular processes, including tumorigenesis, and its overexpression is observed in cell lines and primary patient samples derived from lymphomas, particularly mantle cell lymphoma (MCL). Here we describe the identification and characterization of a potent and selective inhibitor of PRMT5 with antiproliferative effects in both in vitro and in vivo models of MCL. EPZ015666 (GSK3235025) is an orally available inhibitor of PRMT5 enzymatic activity in biochemical assays with a half-maximal inhibitory concentration (IC50) of 22 nM and broad selectivity against a panel of other histone methyltransferases. Treatment of MCL cell lines with EPZ015666 led to inhibition of SmD3 methylation and cell death, with IC50 values in the nanomolar range. Oral dosing with EPZ015666 demonstrated dose-dependent antitumor activity in multiple MCL xenograft models. EPZ015666 represents a validated chemical probe for further study of PRMT5 biology and arginine methylation in cancer and other diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Chung, J. et al. Protein arginine methyltransferase 5 (PRMT5) inhibition induces lymphoma cell death through reactivation of the retinoblastoma tumor suppressor pathway and polycomb repressor complex 2 (PRC2) silencing. J. Biol. Chem. 288, 35534–35547 (2013).
Wang, L., Pal, S. & Sif, S. Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol. Cell. Biol. 28, 6262–6277 (2008).
Pal, S. et al. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 26, 3558–3569 (2007).
Wei, T.-Y.W. et al. Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. Cancer Sci. 103, 1640–1650 (2012).
Powers, M.A., Fay, M.M., Factor, R.E., Welm, A.L. & Ullman, K.S. Protein arginine methyltransferase 5 accelerates tumor growth by arginine methylation of the tumor suppressor programmed cell death 4. Cancer Res. 71, 5579–5587 (2011).
Cho, E.C. et al. Arginine methylation controls growth regulation by E2F1. EMBO J. 31, 1785–1797 (2012).
Pal, S., Vishwanath, S.N., Erdjument-Bromage, H., Tempst, P. & Sif, S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 24, 9630–9645 (2004).
Karkhanis, V., Hu, Y.-J., Baiocchi, R.A., Imbalzano, A.N. & Sif, S. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem. Sci. 36, 633–641 (2011).
Pollack, B.P. et al. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem. 274, 31531–31542 (1999).
Wolf, S.S. The protein arginine methyltransferase family: an update about function, new perspectives and the physiological role in humans. Cell. Mol. Life Sci. 66, 2109–2121 (2009).
Yang, Y. et al. PRMT9 is a Type II methyltransferase that methylates the splicing factor SAP145. Nat. Commun. doi:10.1038/ncomms7428 (2015).
Boisvert, F.-M., Côté, J., Boulanger, M.-C. & Richard, S. A proteomic analysis of arginine-methylated protein complexes. Mol. Cell. Proteomics 2, 1319–1330 (2003).
Dhar, S. et al. Loss of the major Type I arginine methyltransferase PRMT1 causes substrate scavenging by other PRMTs. Sci. Rep. 3, 1311 (2013).
Gonsalvez, G.B. et al. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J. Cell Biol. 178, 733–740 (2007).
Friesen, W.J. et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 21, 8289–8300 (2001).
Meister, G. et al. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 11, 1990–1994 (2001).
Cheng, D. et al. Novel 3,5-Bis(bromohydroxybenzylidene)piperidin-4-ones as coactivator-associated arginine methyltransferase 1 inhibitors: enzyme selectivity and cellular activity. J. Med. Chem. 54, 4928–4932 (2011).
Cheng, D. et al. Small molecule regulators of protein arginine methyltransferases. J. Biol. Chem. 279, 23892–23899 (2004).
Dowden, J. et al. Small molecule inhibitors that discriminate between protein arginine N-methyltransferases PRMT1 and CARM1. Org. Biomol. Chem. 9, 7814–7821 (2011).
Huynh, T. et al. Optimization of pyrazole inhibitors of Coactivator Associated Arginine Methyltransferase 1 (CARM1). Bioorg. Med. Chem. Lett. 19, 2924–2927 (2009).
Purandare, A.V. et al. Pyrazole inhibitors of coactivator associated arginine methyltransferase 1 (CARM1). Bioorg. Med. Chem. Lett. 18, 4438–4441 (2008).
Therrien, E. et al. 1,2-Diamines as inhibitors of co-activator associated arginine methyltransferase 1 (CARM1). Bioorg. Med. Chem. Lett. 19, 6725–6732 (2009).
Wan, H. et al. Benzo[d]imidazole inhibitors of Coactivator Associated Arginine Methyltransferase 1 (CARM1)—Hit to Lead studies. Bioorg. Med. Chem. Lett. 19, 5063–5066 (2009).
Liu, F. et al. Exploiting an allosteric binding site of PRMT3 yields potent and selective inhibitors. J. Med. Chem. 56, 2110–2124 (2013).
Obianyo, O. et al. A chloroacetamidine-based inactivator of protein arginine methyltransferase 1: design, synthesis, and in vitro and in vivo evaluation. ChemBioChem 11, 1219–1223 (2010).
Smil, D. et al. Discovery of a dual PRMT5–PRMT7 inhibitor. ACS Med. Chem. Lett. 10.1021/ml500467h (2015).
Alinari, L. et al. Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation. Blood 10.1182/blood-2014-12-619783 (2015).
Kaniskan, H.Ü. et al. A potent, selective and cell-active allosteric inhibitor of protein arginine methyltransferase 3 (PRMT3). Angew. Chem. Int. Ed. Engl. 10.1002/anie.201412154 (2015).
Copeland, R.A. Evaluation of Enzyme Inhibitors in Drug Discovery: A Guide for Medicinal Chemists and Pharmacologists 2nd edn. (John Wiley & Sons, 2013).
Baell, J.B. & Holloway, G.A. New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem. 53, 2719–2740 (2010).
Sneeringer, C.J. et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl. Acad. Sci. USA 107, 20980–20985 (2010).
Lor, L.A. et al. A simple assay for detection of small-molecule redox activity. J. Biomol. Screen. 12, 881–890 (2007).
Wang, M., Xu, R.-M. & Thompson, P.R. Substrate specificity, processivity, and kinetic mechanism of protein arginine methyltransferase 5. Biochemistry 52, 5430–5440 (2013).
Antonysamy, S. et al. Crystal structure of the human PRMT5:MEP50 complex. Proc. Natl. Acad. Sci. USA 109, 17960–17965 (2012).
Sun, L. et al. Structural insights into protein arginine symmetric dimethylation by PRMT5. Proc. Natl. Acad. Sci. USA 108, 20538–20543 (2011).
Dougherty, D.A. The cation-p interaction. Acc. Chem. Res. 46, 885–893 (2013).
Martinez Molina, D. et al. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87 (2013).
Daigle, S.R. et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20, 53–65 (2011).
Knutson, S.K. et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol. Cancer Ther. 13, 842–854 (2014).
Knutson, S.K. et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 8, 890–896 (2012).
Aggarwal, P. et al. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell 18, 329–340 (2010).
McCurdy, A., Jimenez, L., Stauffer, D.A. & Dougherty, D.A. Biomimetic catalysis of SN2 reactions through cation-.pi. interactions. The role of polarizability in catalysis. J. Am. Chem. Soc. 114, 10314–10321 (1992).
Wasilko, D. Titerless infected-cells preservation and scale-up. BioProcessing Journal 5, 29–32 (2006).
Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010).
Evans, P.R. & Murshudov, G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 (2013).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology: Macromolecular Crystallography, Part A, Vol. 276 (eds Carter, C.W. Jr. & Sweet, R.) Ch. 19 (Academic Press, 1997).
Schüttelkopf, A.W. & van Aalten, D.M.F. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 (2004).
Winn, M.D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 (2011).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 (1997).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Karlsson, R., Katsamba, P.S., Nordin, H., Pol, E. & Myszka, D.G. Analyzing a kinetic titration series using affinity biosensors. Anal. Biochem. 349, 136–147 (2006).
Acknowledgements
We acknowledge team members from Epizyme, Inc. (E.C.-P., K.G.K., C.R.M., P.A.B.-S., T.J.W., L.D.J., N.R., M.J.M., L.J., S.L.J., K.A.W., T.L., K.S., S.A.R., A.R., M.P.S., N.J.W., R.M.P., J.J.S., M.P.M., R.A.C., R.C. and K.W.D.) and GlaxoSmithKline (O.B., M.P., T.F.H., K.N., K.P.O., K.T.G. and R.K.) for their contributions to this manuscript.
Author information
Authors and Affiliations
Contributions
K.W.D., M.J.M. and R.C. designed compounds. L.J. and P.A.B.-S. performed X-ray crystallography and produced X-ray protein. M.P., T.F.H., K.N., K.P.O. and K.T.G. made the initial protein for biochemistry. E.C.-P., K.G.K., T.L. and L.D.J. performed in vitro methyl mark and proliferation studies. N.R. and N.J.W. performed ADME pharmacokinetics studies. E.C.-P., K.G.K., R.M.P. and K.A.W. ran in vivo studies. C.R.M. and T.J.W. ran biochemical and SPR studies. K.W.D., E.C.-P., K.G.K., S.L.J., M.P.S., M.P.M., R.A.C., O.B., R.K., N.J.W., N.R., K.S., J.J.S., R.C., A.R. and S.A.R. designed studies and interpreted results. K.W.D., E.C.-P., K.G.K., T.J.W. and P.A.B.-S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
E.C.-P., K.G.K., C.R.M., P.A.B.-S., T.J.W., L.D.J., N.R., L.J., S.L.J., K.A.W., T.L., K.S., S.A.R., A.R., M.P.S., N.J.W., R.M.P., J.J.S., M.P.M., R.A.C., R.C. and K.W.D. were employees of Epizyme, Inc. at the time of the studies. O.B., M.P., T.F.H., K.N., K.P.O., K.T.G. and R.K. were employees of GSK at the time of the studies. M.J.M. is a consultant for Epizyme, Inc.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1-23, Supplementary Tables 1-3 and Supplementary Note. (PDF 4892 kb)
Rights and permissions
About this article
Cite this article
Chan-Penebre, E., Kuplast, K., Majer, C. et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol 11, 432–437 (2015). https://doi.org/10.1038/nchembio.1810
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1810
This article is cited by
-
PRMT5 inhibition shows in vitro efficacy against H3K27M-altered diffuse midline glioma, but does not extend survival in vivo
Scientific Reports (2024)
-
The ULK3 kinase is a determinant of keratinocyte self-renewal and tumorigenesis targeting the arginine methylome
Nature Communications (2023)
-
Combination treatment of T1-44, a PRMT5 inhibitor with Vactosertib, an inhibitor of TGF-β signaling, inhibits invasion and prolongs survival in a mouse model of pancreatic tumors
Cell Death & Disease (2023)
-
Multimodal perturbation analyses of cyclin-dependent kinases reveal a network of synthetic lethalities associated with cell-cycle regulation and transcriptional regulation
Scientific Reports (2023)
-
Critical Roles of Protein Arginine Methylation in the Central Nervous System
Molecular Neurobiology (2023)