Abstract
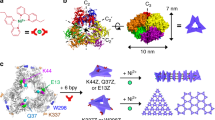
Natural proteins can be versatile building blocks for multimeric, self-assembling structures. Yet, creating protein-based assemblies with specific geometries and chemical properties remains challenging. Highly porous materials represent particularly interesting targets for designed assembly. Here, we utilize a strategy of fusing two natural protein oligomers using a continuous alpha-helical linker to design a novel protein that self assembles into a 750 kDa, 225 Å diameter, cube-shaped cage with large openings into a 130 Å diameter inner cavity. A crystal structure of the cage showed atomic-level agreement with the designed model, while electron microscopy, native mass spectrometry and small angle X-ray scattering revealed alternative assembly forms in solution. These studies show that accurate design of large porous assemblies with specific shapes is feasible, while further specificity improvements will probably require limiting flexibility to select against alternative forms. These results provide a foundation for the design of advanced materials with applications in bionanotechnology, nanomedicine and material sciences.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Whitesides, G. M. & Grzybowski, B. Self-assembly at all scales. Science 295, 2418–2421 (2002).
Goodsell, D. S. & Olson, A. J. Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct. 29, 105–153 (2000).
Lucon, J. et al. Use of the interior cavity of the P22 capsid for site-specific initiation of atom-transfer radical polymerization with high-density cargo loading. Nature Chem. 4, 781–788 (2012).
Douglas, T. & Young, M. Viruses: making friends with old foes. Science 312, 873–875 (2006).
Huard, D. J., Kane, K. M. & Tezcan, F. A. Re-engineering protein interfaces yields copper-inducible ferritin cage assembly. Nature Chem. Biol. 9, 169–176 (2013).
Zhang, S. Fabrication of novel biomaterials through molecular self-assembly. Nature Biotechnol. 21, 1171–1178 (2003).
King, N. P. & Lai, Y. T. Practical approaches to designing novel protein assemblies. Curr. Opin. Struct. Biol. 23, 632–638 (2013)
Lai, Y. T., King, N. P. & Yeates, T. O. Principles for designing ordered protein assemblies. Trends Cell Biol. 22, 653–661 (2012).
Gradisar, H. & Jerala, R. Self-assembled bionanostructures: proteins following the lead of DNA nanostructures. J. Nanobiotechnol. 12, 4 (2014).
Gradisar, H. et al. Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments. Nature Chem. Biol. 9, 362–366 (2013).
Fletcher, J. M. et al. Self-assembling cages from coiled-coil peptide modules. Science 340, 595–599 (2013).
Lanci, C. J. et al. Computational design of a protein crystal. Proc. Natl Acad. Sci. USA 109, 7304–7309 (2012).
Padilla, J. E., Colovos, C. & Yeates, T. O. Nanohedra: using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc. Natl Acad. Sci. USA 98, 2217–2221 (2001).
Sinclair, J. C., Davies, K. M., Venien-Bryan, C. & Noble, M. E. Generation of protein lattices by fusing proteins with matching rotational symmetry. Nature Nanotech. 6, 558–562 (2011).
Lai, Y. T., Cascio, D. & Yeates, T. O. Structure of a 16-nm cage designed by using protein oligomers. Science 336, 1129 (2012).
Lai, Y. T., Tsai, K. L., Sawaya, M. R., Asturias, F. J. & Yeates, T. O. Structure and flexibility of nanoscale protein cages designed by symmetric self-assembly. J. Am. Chem. Soc. 135, 7738–7743 (2013).
King, N. P. et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science 336, 1171–1174 (2012).
King, N. P. et al. Accurate design of co-assembling multi-component protein nanomaterials. Nature 510, 103–108 (2014).
Tozawa, T. et al. Porous organic cages. Nature Mater. 8, 973–978 (2009).
Furukawa, H., Cordova, K. E., O'Keeffe, M. & Yaghi, O. M. The chemistry and applications of metal–organic frameworks. Science 341, 1230444 (2013)
Holst, J. R., Trewin, A. & Cooper, A. I. Porous organic molecules. Nature Chem. 2, 915–920 (2010).
Chen, J. H. & Seeman, N. C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 350, 631–633 (1991).
Pinheiro, A. V., Han, D., Shih, W. M. & Yan, H. Challenges and opportunities for structural DNA nanotechnology. Nature Nanotech. 6, 763–772 (2011).
Shih, W. M., Quispe, J. D. & Joyce, G. F. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature 427, 618–621 (2004).
Iinuma, R. et al. Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-PAINT. Science 344, 65–69 (2014).
Afonin, K. A. et al. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nature Nanotech. 5, 676–682 (2010).
Pavone, V. et al. Crystal structure of an amphiphilic foldamer reveals a 48-mer assembly comprising a hollow truncated octahedron. Nature Commun. 5, 3581 (2014).
Ueno, T. Porous protein crystals as reaction vessels. Chem. Eur. J. 19, 9096–9102 (2013).
Molino, N. M. & Wang, S-W. Caged protein nanoparticles for drug delivery. Curr. Opin. Biotechnol. 28, 75–82 (2014).
Peer, D. et al. Nanocarriers as an emerging platform for cancer therapy. Nature Nanotech. 2, 751–760 (2007).
Walters, M. J. et al. Characterization and crystal structure of Escherichia coli KDPGal aldolase. Bioorg. Med. Chem. 16, 710–720 (2008).
Saul, F. A. Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity. J. Mol. Biol. 335, 595–608 (2004).
Kantardjieff, K. A. & Rupp, B. Matthews coefficient probabilities: improved estimates for unit cell contents of proteins, DNA, and protein–nucleic acid complex crystals. Protein Sci. 12, 1865–1871 (2003).
Rose, P. W. et al. The RCSB Protein Data Bank: new resources for research and education. Nucleic Acids Res. 41, D475–D482 (2013).
Inokuma, Y. et al. X-ray analysis on the nanogram to microgram scale using porous complexes. Nature 495, 461–466 (2013).
Hernandez, H. & Robinson, C. V. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nature Protoc. 2, 715–726 (2007).
Hura, G. L. et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nature Methods 6, 606–612 (2009).
Schneidman-Duhovny, D., Hammel, M. & Sali, A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res. 38, W540–W544 (2010).
Fotin, A. et al. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature 432, 573–579 (2004).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
McCoy, A. J. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997).
Sobott, F., Hernandez, H., McCammon, M. G., Tito, M. A. & Robinson, C. V. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal. Chem. 74, 1402–1407 (2002).
Brunton, A. N., Fraser, G. W., Lees, J. E. & Turcu, I. C. Metrology and modeling of microchannel plate X-ray optics. Appl. Opt. 36, 5461–5470 (1997).
Ebong, I. O. et al. Heterogeneity and dynamics in the assembly of the heat shock protein 90 chaperone complexes. Proc. Natl Acad. Sci. USA 108, 17939–17944 (2011).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Voss, N. R., Yoshioka, C. K., Radermacher, M., Potter, C. S. & Carragher, B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009).
Yang, Z., Fang, J., Chittuluru, J., Asturias, F. J. & Penczek, P. A. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure 20, 237–247 (2012).
Dyer, K. N. et al. In Structural Genomics General Applications (ed. Wai, Y.) Ch. 18, 245–258 (Molecular Methods in Biology Series, Vol. 1091, Springer, 2014).
Acknowledgements
This work was supported by the National Science Foundation (grant CHE-1332907, T.O.Y.), the BER programme of the Department of Energy Office of Science, and the National Institutes of Health (NIH, grant R01GM067167, F.J.A.). The authors thank M. Sawaya, D. Cascio, D. McNamara and D. Leibly for X-ray data collection at the Advanced Photon Source (APS), the staff at APS beamline 24-ID-C and the National Resource for Automated Macromolecular Microscopy (NRAMM) for support. The authors thank D. Woolfson, N. King and members of the D. Baker laboratory for discussions and T. Goddard for advice on modelling in UCSF Chimera. SAXS data collection and analysis at BL12.3.1 at the Advanced Light Source (ALS) was supported by the Integrated Diffraction Analysis Technologies (IDAT) program (DOE/BER), by the Department of Energy (contract DE-AC02-05CH11231) and by NIH MINOS (R01GM105404).
Author information
Authors and Affiliations
Contributions
Y-T.L. and T.O.Y. conceived the project. Y-T.L., E.R., G.H., K-L.T., A.L., J.A.T. and T.O.Y. performed the experiments and analysed the data. Y-T.L. and T.O.Y. drafted the manuscript. All other authors contributed to the writing and editing of individual sections of the complete manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 5540 kb)
Rights and permissions
About this article
Cite this article
Lai, YT., Reading, E., Hura, G. et al. Structure of a designed protein cage that self-assembles into a highly porous cube. Nature Chem 6, 1065–1071 (2014). https://doi.org/10.1038/nchem.2107
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2107
This article is cited by
-
A supramolecular system mimicking the infection process of an enveloped virus through membrane fusion
Scientific Reports (2023)
-
Accurate computational design of three-dimensional protein crystals
Nature Materials (2023)
-
Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials
Nature Communications (2021)
-
Protein-based antigen presentation platforms for nanoparticle vaccines
npj Vaccines (2021)
-
Design of metal-mediated protein assemblies via hydroxamic acid functionalities
Nature Protocols (2021)