Abstract
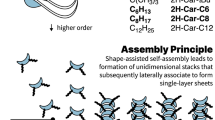
Molecular recognition plays an important role in nature, with perhaps the best known example being the complementarity exhibited by pairs of nucleobases in DNA. Studies of self-assembling and self-organizing systems based on molecular recognition are often performed at the molecular level, however, and any macroscopic implications of these processes are usually far removed from the specific molecular interactions. Here, we demonstrate that well-defined molecular-recognition events can be used to direct the assembly of macroscopic objects into larger aggregated structures. Acrylamide-based gels functionalized with either host (cyclodextrin) rings or small hydrocarbon-group guest moieties were synthesized. Pieces of host and guest gels are shown to adhere to one another through the mutual molecular recognition of the cyclodextrins and hydrocarbon groups on their surfaces. By changing the size and shape of the host and guest units, different gels can be selectively assembled and sorted into distinct macroscopic structures that are on the order of millimetres to centimetres in size.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Cram, D. J. The design of molecular hosts, guests and their complexes. Science 240, 760–767 (1988).
Lehn, J.-M. Supramolecular Chemistry: Concepts and Perspectives (Wiley-VCH, 1995).
Caulder, D. L. & Raymond, K. N. Supermolecules by design. Acc. Chem. Res. 32, 975–982 (1999).
Whitesides, G. M., Mathias, J. P. & Seto, C. T. Molecular self-assembly and nanochemistry: a chemical strategy for the synthesis of nanostructures. Science 254, 1312–1319 (1991).
Fujita, M. et al. Self-assembly of ten molecules into a nanometer-sized organic host frameworks. Nature 378, 469–471 (1995).
Nedelec, F. J., Surrey, T., Maggs, A. C. & Leibler, S. Self-organization of microtubules and motors. Nature 389, 305–308 (1997).
Lehn, J.-M. Toward self-organization and complex matter. Science 295, 2400–2403 (2002).
Sacanna, S., Irvine, W. T. M., Chaikin, P. M. & Pine, D. J. Lock and key colloids. Nature 464, 575–578 (2010).
Harada, A., Li, J. & Kamachi, M. The molecular necklace: a rotaxane containing many threaded α-cyclodextrins. Nature 356, 325–327 (1992).
Hirschberg, J. H. K. K. et al. Helical self-assembled polymers from cooperative stacking of hydrogen-bonded pairs. Nature 407, 167–170 (2000).
Brunsveld, L., Folmer, B. J. B., Meijer, E. W. & Sibesma, R. P. Supramolecular polymers. Chem. Rev. 101, 4071–4098 (2001).
Harada, A., Hashidzume, A., Yamaguchi, H. & Takashima, Y. Polymeric rotaxanes. Chem. Rev. 109, 5974–6023 (2009).
Cordier, P., Tournihac, F., Soulie-Ziakovic, C. & Leibler L. Self-healing and thermoreversible rubber from supramolecular assembly. Nature 451, 977–980 (2008).
Shimizu, T., Masuda, M. & Minamikawa, H. Supramolecular nanotube architectures based on amphiphilic molecules. Chem. Rev. 105, 1401–1443 (2005).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Wasielewski, M. R. Self-assembly strategies for integrating light harvesting and charge separation in artificial photosynthetic systems. Acc. Chem. Res. 42, 1910–1921 (2009).
Rosen, B. M. et al. Dendron-mediated self-assembly, disassembly and self-organization of complex systems. Chem. Rev. 109, 6275–6540 (2009).
Douglas, E. S. et al. Self-assembled cellular microarrays patterned using DNA barcodes. Lab Chip 7, 1442–1448 (2007).
Gartnera, Z. J. & Bertozzi, C. R. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc. Natl Acad. Sci. USA 106, 4606–4610 (2009).
Grzybowski, B. A., Jiang, X., Stone, H. A. & Whitesides, G. M. Dynamic, self-assembled aggregates of magnetized, millimeter-sized objects rotating at the liquid–air interface: macroscopic, two-dimensional classical artificial atoms and molecules. Phys. Rev. E 64, 011603 (2001).
Whitesides, G. M. & Grzybowski, B. Self-assembly at all scales. Science 295, 2418–2421 (2002).
Boncheva, M. et al. Magnetic self-assembly of three-dimensional surfaces from planar sheets. Proc. Natl Acad. Sci. USA 102, 3924–3929 (2005).
Grzybowski, B. A. et al. Electrostatic self-assembly of macroscopic crystals using contact electrification. Nature Mater. 2, 241–245 (2003).
McCarty, L. S., Winkleman, A. & Whitesides, G. M. Electrostatic self-assembly of polystyrene microspheres by using chemically directed contact electrification. Angew. Chem. Int. Ed. 46, 204–209 (2005).
Terfort, A., Bowden, N. & Whitesides, G. M. Three-dimensional self-assembly of millimeter-scale components. Nature 386, 162–164 (1997).
Lahann, J. et al. A reversibly switching surface. Science 299, 371–374 (2003).
Katz, E., Lioubashevsky, O. & Willner, I. Electromechanics of a redox-active rotaxane in a monolayer assembly on an electrode. J. Am. Chem. Soc. 126, 15520–15532 (2004).
Yu, H., Iyoda, T. & Ikeda, T. Photoinduced alignment of nanocylinders by supramolecular cooperative motions. J. Am. Chem. Soc. 128, 11010–11011 (2006).
Borden, N., Terfort, A., Carbeck, J. & Whitesides, G. M. Self-assembly of mesoscale objects into ordered two-dimensional arrays. Science 276, 223–225 (1997).
Kim, E. & Whitesides, G. M. Imbibition and flow of wetting liquids in noncircular capillaries. J. Phys. Chem. B 101, 855–863 (1997).
Choi, I. S., Bowden, N. & Whitesides, G. M. Macroscopic, hierarchical, two-dimensional self-assembly. Angew. Chem. Int. Ed. 38, 3078–3081 (1999).
Pisula, W. et al. From macro- to nanoscopic templating with nanographenes. J. Am. Chem. Soc. 128, 14424–14425 (2006).
Stoddart, J. F. Thither supramolecular chemistry? Nature Chem. 1, 14–15 (2009).
Harada, A., Adachi, H., Kawaguchi, Y. & Kamachi, M. Recognition of alkyl groups on a polymer chain by cyclodextrins. Macromolecules 30, 5181–5182 (1997).
Born, M. & Ritter, H. Side-chain polyrotaxanes with a tandem structure based on cyclodextrins and a polymethacrylate main chain. Angew. Chem. Int. Ed. 34, 309–311 (1995).
Acknowledgements
The authors would like to thank T. Inoue and O. Urakawa (Department of Macromolecular Science, Graduate School of Science, Osaka University) for their help in the measurements of stress and strain of gels. We gratefully acknowledge the financial support from the Japan Science and Technology Agency (JST), the Core Research for Evolutional Science and Technology (CREST) program.
Author information
Authors and Affiliations
Contributions
A. Harada conceived the project and designed the experiments. R.K. contributed to the synthesis of the host and guest gels. A. Harada, Y.T., A. Hashidzume and H.Y. analysed the data and co-wrote the paper. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 817 kb)
Supplementary information
Supplementary information (WMV 943 kb)
Supplementary information
Supplementary information (WMV 1433 kb)
Supplementary information
Supplementary information (WMV 1262 kb)
Supplementary information
Supplementary information (WMV 1911 kb)
Supplementary information
Supplementary information (WMV 1243 kb)
Rights and permissions
About this article
Cite this article
Harada, A., Kobayashi, R., Takashima, Y. et al. Macroscopic self-assembly through molecular recognition. Nature Chem 3, 34–37 (2011). https://doi.org/10.1038/nchem.893
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.893
This article is cited by
-
A 4D printed self-assembling PEGDA microscaffold fabricated by digital light processing for arthroscopic articular cartilage tissue engineering
Progress in Additive Manufacturing (2024)
-
Middle-out methods for spatiotemporal tissue engineering of organoids
Nature Reviews Bioengineering (2023)
-
An approach to MOFaxanes by threading ultralong polymers through metal–organic framework microcrystals
Nature Communications (2023)
-
Pillararene incorporated metal–organic frameworks for supramolecular recognition and selective separation
Nature Communications (2023)
-
Macroscopic supramolecular self-assembly detection based on HALCON machine vision
Journal of Inclusion Phenomena and Macrocyclic Chemistry (2023)